
Comprehensive resistome analysis reveals the prevalence of NDM and mcr-1 in Chinese poultry production
Not since the emergence of enterohemorrhagic E. coli (EHEC) in the 1980s have bacteria gained such attention in the public eye, among food producers, healthcare workers and researchers worldwide. Currently, antimicrobial resistant microorganisms are now recognized as one of the most serious global health threats to both humans and animals worldwide by many national and international agencies alike including the World Health Organization (WHO), the Food and Agricultural Organization of the United Nations (FAO), World Organization for Animal Health (OIE), Centers for Disease Control (CDC), United States Department of Agriculture (USDA), and the Food and Drugs Administration (FDA) etc. These agencies all recognize the importance of antimicrobial resistance control and education in how to stem the flow of antimicrobial resistance in animal production and human health. With the emergence of new antimicrobial resistance genes and traits, the risk of human and animal infection as a consequence of drug resistant microorganisms becomes an ever greater threat to the global population and the food animals it needs as a source of protein.
The recent emergence and first identification of plasmid associated colistin-resistant Enterobacteriaceae in China (1) has garnered significant attention from the science and non-science communities alike because of the recognition that one of the last drugs of resort used to treat human illness has become significantly diminished and may potentially become useless in the near future. But the bigger questions to consider here are … Has the era of antimicrobial drugs for treating so many diseases started to fade? and what is there for the next generation? As the global population soars to 8.5 billion in the next 15–20 years what will be the outcome for human and animal health? Currently, antimicrobial resistant infections causing death worldwide are estimated to be 700,000 people annually in regions of the world where the population income is ranked low to middle (2). While death rates in the US and EU due to antimicrobial-resistant organisms are estimated to be 23,000 and 25,000 respectively (3,4). The recent O’Neill report also adds perspective to the antimicrobial resistance story by estimating that by 2050 drug resistant infections could cost the lives of an individual every 3 seconds or 10 million people (2).
Since the first recognition and documentation of colistin resistance in China (1), there has been significant worldwide attention in regards to assessing where colistin resistance has emerged and where it currently is. In fact, the author of this editorial has also recently published work identifying mcr-1 associated resistance in Avian Pathogenic E. coli (APEC) (5) that cause disease in production birds. This paper is just one of the many papers that have emerged over the last two years reporting the detection and characterization of mcr-associated resistance in animals and humans and their characterization (6-13). As an example of the explosion of information on mcr-1 a Google Scholar® search (as of 05/16/2017) indicates 4,950 hits while a PubMed search in NCBI identifies 266 references.
A better understanding of colistin-associated resistance and its implication for both animal production and human health worldwide are of critical importance in any efforts to combat this emerging resistance. Colistin (polymyxin E) is an old generation drug that was first described in the 1940s; the polymyxins are antibiotic products of Bacillus polymyxa and are peptide antibiotics that act by disruption of cell membrane phospholipids, increasing cell permeability with a detergent like action (14). The drug itself is relatively potent and has systemic toxicity including nephrotoxicity which means its use in humans has been relatively rare until recently. Colistin has been used as an antibiotic in agriculture worldwide but its use has also been variable with China being reported as one of the highest users of colistin (see the following link http://phenomena.nationalgeographic.com/2015/11/21/mcr-gene-colistin/) while other reports indicate that farm use of colistin in some EU countries including Spain, Portugal, France, Estonia and Belgium represents 13, 9, 6, 5 and 3.5% of all farm antibiotic use (see http://sustainablefoodtrust.org/articles/antibiotics/). Other studies report that colistin has been in use in Europe since the 1950s but primarily in pigs for the treatment and prevention of diarrhea in piglets caused by E. coli and Salmonella spp. (see http://www.ema.europa.eu/docs/en_GB/document_library/Report/2013/07/WC500146813.pdf). Indeed, the emergence of colistin resistance and the identification of the colistin-associated resistance gene mcr-1 is likely linked with the use of colistin as a drug in animal production and feed. In an effort to stem the flow of resistance, one of the largest world users China will withdraw approval for the use of colistin as a growth promoter beginning in April 2017, potentially removing more than 8,000 tons of the drug from Chinese farming (15).
In a follow up to mcr-1 detection in China, many other countries and investigators have also assessed their historical and current stocks of isolates for the presence of the mcr-1 gene and its associated phenotypic colistin resistance. To date more than 30 countries worldwide have reported mcr-1 associated resistance in Enterobaceriaceae from many production animals including veal, swine, poultry, cattle etc. (1,5,11,16-18). In addition, many researchers and agencies have conducted prospective and retrospective studies in an attempt to determine when mcr-1 emerged in these countries. In a similar analysis of Chinese poultry isolates, Shen and colleagues (19) noted that mcr-1 was detected in historical isolates dating back to the 1980’s when colistin was first used in China, which would suggest that the timeline for colistin resistance emergence is more than 30 but potentially closer to 40 years ago. These studies would suggest that colistin resistance has probably been around for a long time and its prevalence is probably underestimated. Other countries will need to follow suit to identify potential emergence dates for mcr-1 but also consider removal of colistin from the list of drugs appropriate for animal feed and as a growth promoter or as a treatment if we wish to try and preserve what little strength the drug still has for human health. Of greater concern is that mcr-1 resistance has also been recognized in human disease and the first cases of mcr-1 associated infection were found in humans including a urinary tract infection (20) and clinical cases (21-23). Since this time, clinical cases of infection associated with mcr-1 colistin-resistant organisms have been recognized worldwide.
In a recent study that begins the process of understanding colistin resistance in China, Wang and colleagues [2017] (24) have investigated and comprehensively analyzed the resistome of poultry production in China and its relationship to colistin resistance and carbapenamase-associated resistance linked with the New Delhi metallo-β-lactam associated (NDM-1) gene. With the recognition of NDM and mcr-1 as emerging and very serious threats for the health of production animals and human health, this study is timely and seeks to better understand the movement and origins of resistance in a complicated food system.
In this study, the investigators set out to examine 739 samples collected between November 2014 to August 2015 (when colistin was still in use) from hatcheries, poultry farms, wild birds, humans (farmers), farm dogs, flies, sewage, slaughterhouse samples (ceca and sewage) and retail meat in the Beijing region for the presence of E. coli, Klebsiella and direct analysis of DNA. All samples collected were subjected to DNA isolation for total DNA present and additional samples were plated on CHROMagar to detect Klebsiella pneumoniae (KPC) and MacConkey for detection of E. coli. All DNA samples collected and isolates of E. coli were screened by PCR for genes associated with mcr-1 and carbapenamase (NDM) resistance. Conjugation assays were used to assess the mobility of resistance and whole genome sequencing to assess the chromosome or plasmid location of mcr-1 and NDM. Finally, phylogenetic analysis was applied to construct a tree to estimate the E. coli population structure from the isolates and environments sampled.
The findings of this study are interesting, as the role of the environment, people, birds and flies provide for a complicated system from which to understand how mcr-1 and NDM exist and what are the contributory factors in the emergence and sources of resistance to the system and its various hosts. Of interest, the authors of the study detected high levels of mcr-1 in direct samples from hatcheries, chicken cloacae, ceca and meat ranging from 85–97%. Similarly, direct sample testing (DST) from which DNA was extracted for analysis found higher levels of NDM than E. coli that were isolated ranging from 33–62%, suggesting other organisms present in the DNA were potential sources of these resistance traits indicating that there was vast underestimation of the resistome that was contributing to the resistance that is present. This unknown source of resistance was termed the “phantom resistome”. In each sample set analyzed, recovery rates of resistance for blaNDM and mcr-1 were consistently higher in samples where the whole DNA was examined than where isolates of E. coli recovered from the same sample were screened for the genes at the hatcheries, farms, slaughterhouse, retail products, in farm dogs, the farmers, wild birds and flies.
Further characterization of mcr-1 and NDM resistance traits to determine their genetic location in E. coli isolates recovered found that most of the blaNDM and mcr-1 genes were harbored on plasmids of varying sizes ranging from 3 to 280 kb; however, no co-location of blaNDM or mcr-1 on the same plasmid was evident. Plasmids identified as sources of the mcr-1 included IncX3 and IncX4 types. Conjugation assays were able to demonstrate the mobility of resistance for both the blaNDM and mcr-1 resistance traits among 24 and 20 donors respectively at frequencies of 3.7×10−5 to 3.2×10−1 and 9×10−10 to 5×10−3 cells per donor cell.
Of significant importance in this study is the analysis of sequence types detected among the E. coli strains examined and the demonstration of the association between the sequence types (ST) detected with similar STs found in humans and the food chain. An in-silico multi-locus sequence type analysis (MLST) from the sequenced genomes of carbapenamase-resistant E. coli recovered in the study demonstrated that the same sequence types, ST10 and ST156 could be found in chicken farms, slaughter houses, retail product and humans as well as flies, farm dogs and wild birds. Further analysis using single nucleotide polymorphism analysis (SNP) of the core genome of ST 156 strains found commonality among the STs from the disparate samples suggesting that these strains were being shared or moved around. The study also demonstrates the movement of these resistance traits among poultry, humans, the environment and even international spread of resistance.
One of the important take home messages from this paper is the demonstration of NDM as a dominant mechanism of carbapenem-resistant Enterobacteriaceae (CRE) in China that appears to exist in both animals and humans. Also of concern is the fact that a considerable number of CRE (E. coli) strains sequenced possessed mcr-1 indicating that these organisms were resistant to both carbapenems and colistin and could be detected in a number of locations from the farm to the birds, slaughterhouse, retail products and people. Perhaps the greatest concern that this study highlights is that we still do not fully understand all of the bacterial (and other) reservoirs of these resistance genes and the detection of blaNDM and mcr-1 from direct samples that represented a greater overall prevalence of these two resistance genes than from E. coli alone recovered from these same samples, suggesting other sources of resistance for the “phantom resistome”. These studies therefore suggest we are considerably underestimating the prevalence and levels of carbapenamase and/or mcr-1–associated resistance that is out there. Figure 1 (reprinted with permission) demonstrates the movement of resistance from farms to humans, the food chain and vectors of resistance movement.
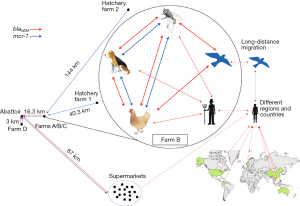
Overall, this paper has done an excellent job in understanding the epidemiology and movement of antimicrobial resistance linked with carbapenamase and colistin in food production in China, but it also raises more questions as to sources of the “phantom resistome” be it bacterial or otherwise and how to approach controlling antimicrobial resistance once it has emerged. Clearly, the study is timely as it describes the current situation with colistin before the withdrawal of the drug and it will be interesting to repeat this study in a few years to determine if the colistin and NDM resistance still remains in the absence of colistin use and its associated pressure.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by Executive Editor Bing Gu (Department of Laboratory Medicine, the Affiliated Hospital of Xuzhou Medical University, Xuzhou, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2017.06.02). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016;16:161-8. [Crossref] [PubMed]
- O'Neill J. Tackling drug-resistant infections globally: final report and recommendations. May 2016.
- Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013;13:1057-98. [Crossref] [PubMed]
- Laxminarayan R, Matsoso P, Pant S, et al. Access to effective antimicrobials: a worldwide challenge. Lancet 2016;387:168-75. [Crossref] [PubMed]
- Lima Barbieri N, Nielsen DW, Wannemuehler Y, et al. mcr-1 identified in Avian Pathogenic Escherichia coli (APEC). PLoS One 2017;12:e0172997 [Crossref] [PubMed]
- Nordmann P, Assouvie L, Prod'Hom G, et al. Screening of plasmid-mediated MCR-1 colistin-resistance from bacteremia. Eur J Clin Microbiol Infect Dis 2016;35:1891-2. [Crossref] [PubMed]
- Haenni M, Metayer V, Gay E, et al. Increasing trends in mcr-1 prevalence among ESBL-producing E. coli in French calves despite decreasing exposure to colistin. Antimicrob Agents Chemother 2016;60:6433-4. [Crossref] [PubMed]
- Teo JQ, Ong RT, Xia E, et al. mcr-1 in multidrug-resistant blaKPC-2 clinical Enterobacteriaceae isolates in Singapore. Antimicrob Agents Chemother 2016;60:6435-7. [Crossref] [PubMed]
- Fernandes MR, McCulloch JA, Vianello MA, et al. First Report of the Globally Disseminated IncX4 Plasmid Carrying the mcr-1 Gene in a Colistin-Resistant Escherichia coli Sequence Type 101 Isolate from a Human Infection in Brazil. Antimicrob Agents Chemother 2016;60:6415-7. [Crossref] [PubMed]
- Irrgang A, Roschanski N, Tenhagen BA, et al. Prevalence of mcr-1 in E. coli from Livestock and Food in Germany, 2010-2015. PLoS One 2016;11:e0159863 [Crossref] [PubMed]
- Veldman K, van Essen-Zandbergen A, Rapallini M, et al. Location of colistin resistance gene mcr-1 in Enterobacteriaceae from livestock and meat. J Antimicrob Chemother 2016;71:2340-2. [Crossref] [PubMed]
- El Garch F, Sauget M, Hocquet D, et al. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin Microbiol Infect 2017;23:51.e1-51.e4. [Crossref] [PubMed]
- Doumith M, Godbole G, Ashton P, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 2016;71:2300-5. [Crossref] [PubMed]
- Prescott JF. Peptide Antibiotics: Polymyxins, Glycopeptides, Streptogramins, and Bactiracin. In: Prescott JF, Baggot JD, Walker RD. editors. Antimicrobial Therapy in Veterinary Medicine, 3rd Edition. Ames: Iowa State Press, 2000:177-90.
- Walsh TR, Wu Y. China bans colistin as a feed additive for animals. Lancet Infect Dis 2016;16:1102-3. [Crossref] [PubMed]
- Hasman H, Hammerum AM, Hansen F, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 2015;20: [Crossref] [PubMed]
- Grami R, Mansour W, Mehri W, et al. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill 2016;21:30144. [Crossref] [PubMed]
- Perrin-Guyomard A, Bruneau M, Houee P, et al. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill 2016;21: [Crossref] [PubMed]
- Shen Z, Wang Y, Shen Y, et al. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 2016;16:293. [Crossref] [PubMed]
- McGann P, Snesrud E, Maybank R, et al. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First report of mcr-1 in the USA. Antimicrob Agents Chemother 2016;60:4420-1. [Crossref] [PubMed]
- Castanheira M, Griffin MA, Deshpande LM, et al. Detection of mcr-1 among Escherichia coli Clinical Isolates Collected Worldwide as Part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother 2016;60:5623-4. [Crossref] [PubMed]
- Corbella M, Mariani B, Ferrari C, et al. Three cases of mcr-1-positive colistin-resistant Escherichia coli bloodstream infections in Italy, August 2016 to January 2017. Euro Surveill 2017;22: [Crossref] [PubMed]
- Vasquez AM, Montero N, Laughlin M, et al. Investigation of Escherichia coli Harboring the mcr-1 Resistance Gene - Connecticut, 2016. MMWR Morb Mortal Wkly Rep 2016;65:979-80. [Crossref] [PubMed]
- Wang Y, Zhang R, Li J, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2017;2:16260. [Crossref] [PubMed]
Cite this article as: Logue CM. Comprehensive resistome analysis reveals the prevalence of NDM and mcr-1 in Chinese poultry production. J Lab Precis Med 2017;2:29.


