
Tumor aneuploidy correlates with hallmarks of cancer
Abnormalities of cellular DNA content have long been associated with tumorigenesis. Such abnormalities were originally implicated in cancer development over 100 years ago by German biologist Boveri (1), and during the last years it has become clear that cellular aneuploidy is a driving force in the process of carcinogenesis. One of the major works demonstrating this was published by Davoli et al. in 2013 (2), and in a recent work (3), Davoli et al. illuminate this further by showing that tumor aneuploidy correlates with hallmarks of cancer.
Caspersson (4) was the first author to demonstrate an objective in situ quantification of DNA. He used stoichiometric DNA staining to enable quantitative measurement of the DNA content of individual cell nuclei in a cell sample, and his DNA-content histogram method continues to be used as the accepted approach to describe the population frequencies of nuclei containing different quantities of DNA in the context of the cell cycle of diploid cells (the modern versions of this method are flow cytometry and image cytometry). Since the 1960s, karyotyping has allowed for ploidy determination as well as the detailed chromosome analysis of cells in metaphase after in vitro culture of tissue samples. The resolution of copy number detections increased greatly with newer methods such as arrays and sequencing, but the main downside of this higher resolution was that copy number was no longer obtained per cancer cell, but rather as a relative number compared to a reference state.
However, this changed with the introduction of computational methods that estimate tumor purity (fraction of abnormal vs. normal cells in the sample) and malignant cell ploidy directly from analysis of somatic DNA alterations. In particular, in 2012, the ABSOLUTE method was introduced (5). This method allows the use of additional information from pre-computed statistical models of recurrence cancer karyotypes and somatic point mutations.
The study by Davoli et al. presented in Science earlier this year (3) utilizes this ABSOLUTE method while addressing the complex interplay between distinct types of copy number alterations and cell proliferation and immune evasion, two hallmarks of cancer. While certain focal aberrations, such as a deletion leaving a single mutated allele of a tumor suppressor, may be identified as likely driver events in tumor development, the relation between chromosomal instability and the hallmarks of cancers is, in general, far from being well understood. Several factors contribute to the challenges of studying aneuploidy and its effects. Gains and losses typically affect large numbers of potentially influential genes, aberrations interact with mutations in their effects on tumor development and intratumor heterogeneity may add substantially to the complexity. Moreover, tumor samples are typically analyzed at a fairly late stage in tumor development, and gains or losses that act as driver events are potentially accompanied by large numbers of passenger alterations. Finally, the commonly observed event of genome doubling may, in itself, have limited effects on cell vitality, but is likely to contribute substantially to subsequent instability and clonal evolution by allowing aberrations to occur in ways that maintain wildtype alleles, which protect the cell from apoptosis.
The article by Davoli et al. brings us one step closer to understanding how different forms of tumor aneuploidy relates to cancer development. Their analyses are based on 5,255 tumor/normal samples from 12 cancer types retrieved from the Cancer Genome Atlas project. The analyses focus on two hallmarks of cancer: increased cell proliferation and immune evasion. Since aberrations may either be focal, only affecting few genes, or include whole arms/chromosomes (or even the complete genome), Davoli et al. relates their findings on immune evasion and proliferation to these main types of aneuploidy. Moreover, they address the important interplay between aberrations and mutations.
The use of data from 12 cancer types from the Cancer Genome Project ensures a firm statistical basis for conclusions and enables Davoli et al. to address questions on the generality of the relations revealed. There are certain costs of using such large data sets collected internationally in terms of lack of detailed knowledge of the data and also in terms of the possibility for the investigators to adapt the data collection process to specific biological challenges. However, given that data on aberrations are noisy and for the purpose of revealing to what extent effects are general or cancer type specific, the use of such large data sets will often be vital.
Davoli et al. first examine the relation between aberration level and other genetic features of cancer, particularly mutations. The relation between mutations and copy number alterations appears clearly cancer type dependent. For 8 of the 12 examined cancer types, the correlation is positive, although only strong for breast cancer. Significantly negative correlation was only seen for colorectal cancer and endometrial carcinoma. Interestingly, in these two latter cases the negative correlation is caused by a limited number of “hypermutated” cases with a roughly tenfold increase in number of mutations compared to the rest of the tumors. Davoli et al. also examines the relation between aneuploidy and mutations in specific genes known as potential cancer drivers, and e.g., show that genes involved in the DNA damage response pathway were clearly positively correlated to the level of copy number changes.
Davoli et al. convincingly demonstrate that high levels of copy number changes are correlated to an increased proliferation rate, as revealed by elevated expression of cell cycle and cell proliferation markers. This relation is seen both in the pan-cancer analysis and for all cancer types except colorectal cancer. Sustained proliferation was especially connected to focal aberrations, although also seen for copy number changes at arm/chromosome level. A connection to focal aberrations indicates a mechanism related to the action of specific genes targeted by these aberrations. It should be pointed out that the relation between aberration level and proliferation is a statistical one, there are substantial numbers of tumors with high aberration level without increased level of proliferation markers. However, this is to be expected if specific genes are involved, given that in many of the examined samples there may be large numbers of passenger aberrations not affecting these vital genes.
Much of the detailed analysis carried out by Davoli et al. relates to immune evasion. This is rational, given that such information may potentially be used to improve immune checkpoint blockade, which is a promising therapy for certain cancer types. In statistical terms, there was a strong relation between aberration level and down-regulation of gene expression signatures associated with the immune system. Notably, markers expressed by CD8+ cells and NK cells were markedly reduced in tumors with high levels of copy number alterations. The relation was observed both in the pan-cancer analysis and also for all cancer types with the unsurprising exception of brain tumors. Davoli et al. found that immune evasion seemed especially related to copy number changes at the arm/chromosome level, and from this they conclude that the observations on immune evasion are consistent with a mechanism related to imbalance in gene dosage rather than the action of specific genes.
To address the potential clinical relevance of the findings on immune evasion, Davoli et al. further analyze data from two published clinical trials of immunotherapy (CTLA-4 blockade) in melanoma patients. High levels of copy number alterations were associated with a poorer prognosis, with roughly a doubling of the hazard ratio for those with aberration level above the median compared to those below. Similar relations between high number of copy number changes and poor survival have also been seen in other studies (6-8), although not consistently (9,10). The lack of consistency is perhaps not surprising, since while copy number changes selected for in the cancer evolution process must be expected to confer growth advantage, disordered genomic organization will in general often decrease viability by inducing defects in the mitotic process, for instance by affecting chromosomal segregation or spindle assembly. Davoli et al. also examined the effect of the level of mutations on survival; this relation was less convincing, but strong enough to suggest that a combination of aberration and mutation level might provide a useful biomarker for prediction of survival following the immunotherapy.
The methods used by Davoli et al. include estimation of “tumor purity” as a step in the determination of calls for copy number changes. The ABSOLUTE method is used for most cancer types (otherwise pathology reports are utilized). Such methods necessarily depend on sets of assumptions, and the study of Davoli et al. among others depends on an assumed average ploidy of solid tumors of 3N derived from data in the Mitelman database. Such estimates may be questioned, as karyotype analysis is carried out on only those cells that happen to divide in vitro, and the possible inaccuracies or simplifications implied by the assumptions add to the noise level in the analysis. However, as we assess it, these methodological challenges probably have a limited effect in terms of inducing systematic bias in the study of Davoli et al., and thus do not challenge the main conclusions on proliferation and immune evasion.
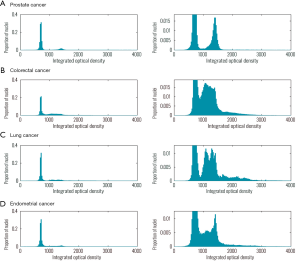
However, although not directly related to the primary questions addressed by Davoli et al., one may question whether the use of the term “tumor purity” is suitable when communicating tumor biology. A tumor is a biological entity evolving through the complex interplay between the genetically altered and the genetically normal cells, the supportive tissue including blood vessels and the opposing forces from the immune system. The expression “tumor purity” restricts the term “tumor” to only the genetically altered malignant cells, focusing on a restricted part of the evolving biological entity. Modern biotechnology provides detailed views into the genomic part of tumor development—and by doing this obviously provides extremely relevant information—but it is important to prevent this leading towards a neglect of a system level understanding of tumor development. In this context, and in relation to the general discussion on copy number alterations in tumors, it may be important to have an appropriate picture of the level of aneuploidy typically observed in tumor samples from different cancer types. Therefore we have calculated DNA content histograms from a large number of tumor samples from prostate cancer, colorectal cancer, lung cancer and endometrial cancer to illustrate the proportions of aneuploid and diploid tumor cells within tumors as well as to illustrate the differences between cancer types (Figure 1). DNA content is estimated from cell suspensions of isolated cell nuclei prepared from formalin fixed and paraffin embedded tissue specimens and assessed with image cytometry (12). A histogram of DNA content estimates is used to illustrate the result. With the exception of lung cancer (biopsies), these samples are analyzed from tumor regions microdissected by pathologist with emphasis on optimizing the number of epithelial cancer cells. The frequency of cell populations with gross genomic aberrations differs significantly between cancer types. While most colorectal cancers have cell populations with a non-diploid DNA content, this is far less common in prostate cancers. For all the illustrated cancer types, however, the majority of cells in the cancer have a diploid DNA content (average proportion: 69%), underlining the importance of cell populations without gross genomic aberrations.
Theodor Boveri was remarkably accurate in his predictions about chromosomal instability as a key hallmark of cancer 100 years ago. Despite the early awareness of the importance of aneuploidy in cancer and numerous studies demonstrating the prognostic relevance, only limited knowledge has been gained about the relationships between aneuploidy and other key properties of carcinogenesis. The study by Davoli et al. is an important contribution and indicates one of the main directions of this research field in the time to come.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Fang Wang (Department of Laboratory Medicine, the First Affiliated Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2017.07.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boveri T. Concerning the Origin of Malignant Tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 2008;121:1-84. [Crossref] [PubMed]
- Davoli T, Xu AW, Mengwasser KE, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013;155:948-62. [Crossref] [PubMed]
- Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355: [Crossref] [PubMed]
- Caspersson TO. History of the development of cytophotometry from 1935 to the present. Anal Quant Cytol Histol 1987;9:2-6. [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Carter SL, Eklund AC, Kohane IS, et al. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043-8. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Hveem TS, Merok MA, Pretorius ME, et al. Prognostic impact of genomic instability in colorectal cancer. Br J Cancer 2014;110:2159-64. [Crossref] [PubMed]
- Baumbusch LO, Helland A, Wang Y, et al. High levels of genomic aberrations in serous ovarian cancers are associated with better survival. PLoS One 2013;8:e54356 [Crossref] [PubMed]
- Jamal-Hanjani M, A'Hern R, Birkbak NJ, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann Oncol 2015;26:1340-6. [Crossref] [PubMed]
- Hveem TS, Kleppe A, Vlatkovic L, et al. Chromatin changes predict recurrence after radical prostatectomy. Br J Cancer 2016;114:1243-50. [Crossref] [PubMed]
- Pradhan M, Abeler VM, Danielsen HE, et al. Image cytometry DNA ploidy correlates with histological subtypes in endometrial carcinomas. Mod Pathol 2006;19:1227-35. [Crossref] [PubMed]
Cite this article as: Danielsen HE, Hveem TS, Liestøl K. Tumor aneuploidy correlates with hallmarks of cancer. J Lab Precis Med 2017;2:51.


