
Integration of ISO 15189 and external quality assurance data to assist the detection of poor laboratory performance in NSW, Australia
Introduction
In Australia, the evaluation of pathology laboratory performance is primarily achieved through conformance assessment to ISO 15189 and National Pathology Accreditation Advisory Council (NPAAC) standards and external quality assurance (EQA) processes. Accreditation and proficiency testing/EQA (PT/EQA) processes provide distinct monitoring methods to ensure ongoing excellence for pathology laboratory quality. The ISO 15189 and NPAAC standards assessing body is the National Association of Testing Authorities (NATA) which uses regular laboratory audits and standard non-conformances ranging from no comments (situation satisfactory), observations, minor infractions that require attention, to conditions that must be met within a given timeframe to ensure ongoing accreditation (1). The EQA method of quality evaluation is conducted by the Royal College of Pathologists of Australasia Quality Assurance Programs (RCPAQAP) and involves achieving acceptable performance against defined analytical performance specifications (APS). Individual assay results from individual laboratories are assessed against the known target value and APS, and unacceptable performance flagged back to the laboratory. These results form part of the ISO 15189 audit and are used to provide a measure of quality performance.
The identification of poorly performing pathology laboratories is a major task of health care regulators. The vast majority of laboratories provide timely and accurate results to referring clinicians, but some laboratories are a danger to patients because of poor clinical governance and/or supervision and poor quality control of assays or inadequate training of staff. Finding out that a laboratory is a poor performer relies on complaints by referrers, often at a stage when there has been potentially significant patient impact; or on some form of inspection, either routine or random, with or without phantom samples. All Australian laboratories are required to be in an EQA program (PT), routinely analyse test samples and report all assays that they report on patients. It would be beneficial to regulators and patients tempting if the results of these samples could be used as a flag to identify potentially poorly performing laboratories.
Combining these two sources of laboratory performance data potentially will enhance the ability to detect quality control problems with greater accuracy and efficiency, achieved by cross-referencing EQA data back to ISO 15189 audit results, and vice versa, thus allowing both evaluation process to inform the other on performance decline detected via their specific metric. Combining data sources produces a larger data set with linked inputs (EQA data) and outputs (ISO 15189 audit data), enabling predictions to be made, increasing the accuracy of predictions emanating from the models.
A systematic survey (SS) of the biomedical literature pertaining to pathology laboratory quality performance was conducted, with reference to “external quality assurance” processes and standards such as ISO 15189. Sample ISO 15189 audit and EQA data collected from NSW laboratories over 2015 were also interrogated via machine learning and linear models to complement the SS study. Past international results lacked consistency or consensus concerning the measurement of laboratory quality reporting data, but a sub-section of reports suggested root cause analysis (RCA) as a suitable method to detect and monitor performance. Therefore, a unified model of quality performance assessment, using data collected as separate external quality control processes (ISO 15189 audit and EQA), was the ultimate aim of this study.
Materials and methods
Systematic survey
To understand best-practice internationally, a SS of the quality literature was performed via PubMed (nlm.nih.gov). Key search (MeSH) terms were: “15189 OR ISO 15189”, “proficiency testing”, “pathology laboratory performance”, and “external quality assurance”. From the total collection of articles identified in PubMed, papers were further sub-divided into articles that analysed quality for the core laboratory functions, namely routine chemistry and haematology, while papers on histology, cytology and specialist laboratories, for example, molecular pathology, immunology, coagulation studies, were excluded from the primary analyses. There were many articles reporting the successes and challenges at a country or regional level, which were also sub-grouped for dedicated analysis. Peer-reviewed articles identified for this pilot study had date ranges from 1992 to 2016 (Figure 1). Articles from journals not available via the authors’ institutional library electronically or in hardcopy were obtained via “Article Reach”, and inter-university document delivery services (www.anu.edu.au/library).

Text-mining analyses were performed on 103 papers reporting on results and/or review of pathology quality for core laboratories, then identical analyses were performed on sub-groupings within this collection, namely, papers that only discussed “EQA” or “15189”, as well as articles that reported on investigations of national systems (Figure 1). (Articles reporting national results were collected for: Belgium, Croatia, Italy (including regions), Jordan and the Middle-East in general, Burkina Faso, Ghana, Korea, Malaysia, Indonesia, the Netherlands and the United States. There is a large literature on this topic from Japan, but all were published in Japanese. Papers published in English, French and Mandarin were included.
Supplementary materials provide the complete list of publications extracted for this study. Additional searches targeted “ISO15189 (AND) EQA” papers, as well as “ISO15189 (NOT) EQA”, and other variations as reported therein. It must also be noted that the ISO15189 standard was introduced internationally in 2003, but not all countries abide by this standard, hence making analysis of trends from the literature difficult. A summary of the final search strategy is shown in Figure 1, which covers the periods pre- and post- the introduction of ISO15189 [1992–2016].
Laboratory quality data
Anonymous, non-identifiable NATA and RCPAQAP data were provided by the NSW Health Pathology (Chatswood, NSW). A sample from 21 laboratories was used for this study, which comprised two laboratory categories, B and G (see below).
For the Australian regulatory framework, laboratories are classified based on the medical governance structure. A category G laboratory is defined to be a laboratory, or a number of co-located laboratories, performing services in one or more groups of pathology testing: (I) under the full-time supervision and clinical governance of a designated person who must be a pathologist, and (II) where (responsibility for supervision of pathology testing may be delegated to other pathologists with relevant scope of practice. These pathologists may further delegate supervision of specific testing to Clinical Scientists with the relevant scope of practice. A category B laboratory is a laboratory performing services in one or more groups of pathology testing, being a laboratory related to an accredited category G laboratory. B laboratories are branch laboratories under the direction and control of a parent G laboratory.
Data—ISO 15189 audit
NATA inspection results for B or G category NSW laboratories reported on “Management Requirements” (ISO 15189 clauses 4.1–4.15, including sub-clauses) and “Technical Requirements” (ISO 15189 clauses 5.1–5.10, including sub-clauses). All ISO 15189 audit reports assess clinical chemistry compliance, as well as haematology and a mixture of other routine and special disciplines. Independent of the additional disciplines besides clinical chemistry, the total laboratory performance was quantitated for analysis.
Non-conformance against ISO 15189 and NPAAC standards were identified by the number of “M” and “C” recommendations recorded for each laboratory. We can do this because against each of the ISO 15189 clauses a blank space, “observation” (O—for noting only), “minor condition” (M—minor non-compliance) or “condition” (C—major non-compliance or condition) was recorded. Since the detection and prediction of poor performance were our aims, the number of M and C observations per laboratory were tallied; C indicates a “condition” associated with poor practice that must be addressed with evidence in the recommended time frame, or an infringement recorded, while M indicates a problem that if not addressed could lead to an upgrade to C classification, and the risk of negative consequences.
M and C categories were derived prior to statistical investigation, for the entire sample of 21 laboratories. C category [0] comprised laboratories with ≤5 total condition reports in total (range, 0–5 reports; median =2), with category [1] represented by labs with ≥7 reports (range, (7–15 reports; median =8). M categories were similarly assigned; M category [0] ranged from 2–7 reports (median =4.5), and M category [1] from 8–18 reports (median =10) of minor conditions for attention by management. These ISO 15189 audit minor (M) and condition (C) categories were used for all subsequent Random Forest (RF) and ANCOVA modelling with EQA results. The number of C or M counts were tallied across all management and technical ISO 15189 audit criteria.
Data—EQA
EQA data was represented by 16 RCPAQAP assessment rounds over 2015 calendar year, performed on the same (21 labs as assessed via NATA ISO 15189 audit.
The serum/blood markers chosen for analysis in this project were: alanine aminotransferase (ALT), aspartate aminotransferase (AST), bicarbonate, total bilirubin, serum calcium, chloride, creatinine kinase (CK), serum creatinine, gamma glutamyl transferase (GGT), serum magnesium, serum phosphate, serum K+, total protein (TP), and serum sodium. Each of these assays did not vary by more than two standard deviations across the automated platforms employed by NSW pathology laboratories, thus eliminating platform-associated measurement variation as a factor in the data modelling and analyses.
Bias was used as an indicator of variation to gauge EQA performance for selected markers, and by extension pathology laboratories. Bias estimates assay accuracy through calculating the difference between the individual laboratory result, and the target values as set by the RCPAQAP. Mean or median bias was calculated for all laboratories (n=21), as well as separately for B (n=10) and G (n=11) labs, and used as the target value for analysis for all of the serum markers listed. The final results were thereafter expressed as a percentage of the target value.
Statistics and text mining
Text mining
Text mining was conducted on groups/sub-groups within the assembled publication collection, using the R (version 3.3.1) (statistical language (1). R packages employed were, tm, SnowballC, ggplot2, wordcloud, and cluster (2-7). All articles were saved as PDF documents prior to uploading into R, and analysis. Word frequency, word clouds and dendrograms were constructed to ascertain dominant word patterns in the selected texts, supported by word correlation results.
Random Forests of combined ISO 15189 audit/EQA results
RF were conducted via R (version 3.3.1) statistical programing (1), using the package randomForest (8,9). RF were run with 1–3 trees per cycle, on 10,000 trees in total. Because of small numbers, a bootstrapping function was added to enhance accuracy via sampling with replacement. Accuracy of M or C category prediction by percentage bias was calculated as an “out-of-bag” (OOB) estimate of success in predicting the correct M or C category. Due to small sample numbers, no other machine learning algorithms were applied.
Analyses of ISO 15189 audit
Total counts of NATA auditor assigned “Conditions” (C) and “Minor” (M) observations recorded for each laboratory, as an assessment of management and technical performance, were analysed by non-parametric statistical methods (SPSS, version 22) (10). To understand variation in C and M counts between G and B laboratories, Kruskal-Wallis tests were performed, and for the investigation of whether C and M counts varied significantly for NATA management versus technical criteria, the Friedman test was conducted. For both non-parametric analyses, significance was set at P<0.05.
ANCOVA of integrated ISO 15189 audit plus EQA covariates
To understand the effect on bias of NATA ratings, other EQA measurements of bias and laboratory type (B or G), analysis of co-variance (ANCOVA) was conducted for the 21 NSW pathology laboratories in the study sample. GGT (%) (bias was the dependent variable, with laboratory type added to the model as a fixed factor, and ISO 15189 audit condition (C) or minor (M) counts (for each laboratory), as well as EQA (%) bias results included as covariates. Analyses were performed using SPSS (version 22.0) (10) as a general linear model. With the addition of covariates, a regression modelling (main effects) was also possible in addition to standard ANOVA outputs (type III sum-of-squares). In addition to significance at P<0.05, effect size was calculated as partial Eta-square (Eta2). GGT (%) bias was selected as a representative EQA marker due to its role as the lead predictor of NATA M category.
Results
The PubMed literature search using the aforementioned search terms yielded 144 primary manuscripts, with full manuscripts in PDF obtained for all but 6 of the total manuscripts identified. The final text analysis was applied to 103 articles, with 37 excluded because of a focus on specialised laboratory functions, for example, quality for coagulation assays, molecular pathology, or advanced immune or microbiological functions (Figure 1).
Text mining
Word frequency
Figure 2 summarises the results of text mining analyses via wordcloud and word frequency involving sub-groupings of articles based on EQA or ISO 15189 focus, country focus, and key words identified from analyses of all texts. Text mining of all 103 articles showed similar word patterns and frequencies.
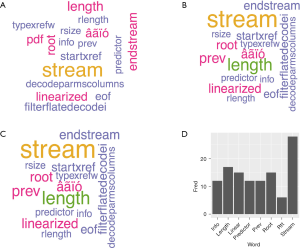
“Stream” was the most frequent word detected, but represented several words/terms, namely, “downstream”, “stream lining” and “stream mapping” that were not consistent. Nonsense words, for example, “âãïó”, “eof”, “fontdescriptor”, were identified as PDF-associated code required to produce the document, and were eliminated from the analysis. Following “stream” in prevalence were “root”, “linear” or “linearized”, and “predict” or “prediction”, reflected also by cluster dendrogram results (Figure 3). The strongest cluster (closest to the Y-axis) featured “predictor” and “info” as associated, with “root” and “linearized” located in the next strongest word cluster. Closer examination via word searches on individual articles found that part-words like “prev” and “info” were due to a number of larger words (e.g., “preview”, “previous”), and as such no useful word patterns were found. “Predictor” and “linearized” were detected regularly in this literature (e.g., “predictor” data “linearized” by square root to fit statistical models). “Root cause” emerged as relevant to the assessment of pathology laboratory quality, with 13 articles identified that discussed this concept. References to “root cause” were linked to the identification of quality failure, and to analyses to uncover systematic failures in quality control (11-14).

Text correlation
The calculation of word correlations was also available via R text mining algorithms. Examination of correlation for the analytical terms “predictor” and “linearized” found a strong association (r=0.57). “Predictor” and “root” (as in square root, root cause) had an identical correlation of r=0.57. “Linearized” and “root” had a perfect correlation (r=1.00), supported by cluster analysis (Figure 3). Correlation between the three key words “predictor”, “root” and “linearize” to other frequent words detected (Figure 2) had moderate to poor associations ranging from 0.0<r<0.4.
Integration of ISO 15189 audit and RCPAQAP data
Comparison of ISO 15189 audit performance categories
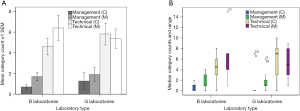
Figure 4 summarises the number of M and C reports across ISO 15189 audit management and technical performance, for B and G laboratories during 2015. These data are presented as: (I) mean ± standard error of the mean (SEM) and, (II) median with interquartile ranges. There were no significant differences overall (management and technical M and C frequency) when comparing B and G laboratories (Kruskal-Wallis, P=0.17–0.72). However, a significant difference was detected for the frequency of M and C counts when comparing management versus technical performance, with technical counts significantly higher than management (P<0.001; Freidman test, df=3; χ2=37.15). In light of this result, a stronger focus on technical competence within laboratories is suggested in relation to quality assessment.
RF predictions of ISO 15189 audit performance by EQA program data
Data used for the RF modelling comprised: total (%) bias for each EQA assay/marker and counts of ISO 15189 audit (M) and (C) categories for laboratory management and technical performance over 2015. The number of M and C comments reported by NATA auditors for each laboratory were recorded, the median calculated for each class, and laboratories classified as above [1] or below [2] the group median. This simple classification allows laboratories to benchmark their achievement, and simplifies the statistical modelling in the face of the small sample size, as well as small variation in the number of M and C comments recorded (Table 1).
Table 1
| Between-subjects effects for GGT (%) bias (DV) | Type III sum of squares | df | Mean square | F | Sig. | Partial Eta2 | Observed powerb |
|---|---|---|---|---|---|---|---|
| Corrected model | 1080.14a | 4 | 270.04 | 30.20 | 0.000 | 0.88 | 1.000 |
| Intercept | 138.20 | 1 | 138.20 | 15.45 | 0.001 | 0.49 | 0.960 |
| Lab category (B or G) | 22.31 | 1 | 22.31 | 2.50 | 0.134 | 0.14 | 0.320 |
| Bicarbonate (%) bias | 9.81 | 1 | 9.81 | 1.10 | 0.310 | 0.06 | 0.170 |
| K+ (%) bias | 243.89 | 1 | 243.89 | 27.27 | <0.001 | 0.63 | 0.998 |
| Total NATA M count* | 73.56 | 1 | 73.56 | 8.23 | 0.011 | 0.34 | 0.770 |
| Error | 143.08 | 16 | 8.94 | – | – | – | – |
| Total | 6812.50 | 21 | – | – | – | – | – |
| Corrected total | 1223.23 | 20 | – | – | – | – | – |
*, management + technical minor reports; a, R2 =0.883 (adjusted R2 =0.854); b, computed using alpha =0.05; GGT, gamma glutamyl transferase; EQA, external quality assurance; NATA, National Association of Testing Authorities; DV, dependent variable; Sig., significance.
The following RF analyses (RFA) rank the importance of individual RCPAQAP assays as predictors of high or low M and C counts, as reflected by the high/low categories described. The results (Figure 5) also present an estimate of error rate (%) in the prediction of M and C category prediction by RCPAQAP data.
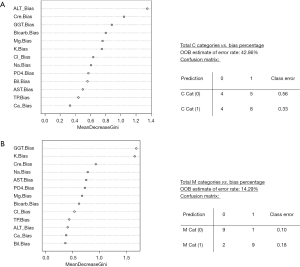
Figure 5 summarises the RFA results interrogating the question of which RCPAQAP assay profiles most accurately predict ISO 15189 audit non-conformance (C, Figure 5A) or (M, Figure 5B) categories. This represents the percentage (%) bias calculated for each assay (ALT etc.) across all (21 NSW laboratories (due to small sample size, a B versus G laboratory category analysis was not possible). Three bias categories (0= top 20%, 1= middle 20–90%, 2= bottom >90%) were also tried versus ISO 15189 audit C and M categories.
The results of the RFA included the percentage accuracy in terms of predicting the ISO 15189 audit C or M categories via the RCPAQAP (%) bias results, as calculated as an OOB estimate, and a “confusion matrix” that reports the number of correct/incorrect cases predicted by RFA per C or M category (Figure 5). Figure 5A summarises the RF prediction of C category by RCPAQAP (%) bias results. An overall error rate of 43% (42.86%) was calculated by the model, indicating successful predictions at 57% based on the RCPAQAP assay result pattern led by ALT and serum creatinine. For this RF model the prediction of C category [0] was poor, with 56% (0.56) of cases incorrectly predicted (while the correct prediction of C category [1] was superior at 67%).
The results for C category prediction stand in contrast with the identical RFA for M category prediction via RCPAQAP (%) bias results. The overall model (Figure 5B) (recorded an OOB error of 14% (14.29%), conversely indicating an overall model accuracy of 86%. Inspection of the confusion matrix shows a prediction accuracy of 90% (0.10 error) for M category [0], and 82% (0.18 error) accuracy for the prediction M category [1]. The leading RCPAQAP predictors for the M category RFA were GGT and serum K+ (mean Gini decrease of >1.5), followed by serum creatinine (mean Gini decrease of ~1.0). Interestingly, ALT was the leading EQA predictor for the C category model.
RCPAQAP assay bias and ISO 15189 audit results modelling
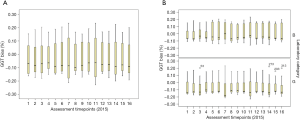
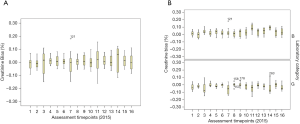
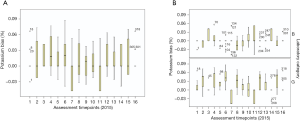
Figure 6 summarises the bias variation of a sample of pathology assay markers from the 2015 RCPAQAP, while Figures 7-9 explore the variation for the 16 QAP-prescribed 2015 time points, as observed for GGT, serum creatinine and K+. The choice of these three specific markers were informed by RF results (Figure 5), and Figure 4.
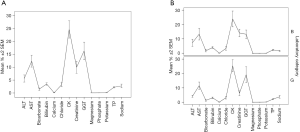
The overall bias across 21 laboratories was highest for the enzymes CK, GGT and AST (but not ALT), with the serum electrolytes and TP below a relative bias value of 5 (Figure 6). In the comparison between B and G laboratory categories, the bias pattern was identical except for serum creatinine, with B category bias greater than 15%, compared to mean G laboratory bias of less than 10%. This observation suggests that serum creatinine is potentially useful for detecting quality deficiencies for B category laboratories, as well as separating B versus G laboratory RCPAQAP performance.
The rates of total C and M scores for B laboratories compared to G laboratories were not significantly different (Figure 4; P>0.16), while the rate of technical category C and M comments were significantly greater than C and M for the management category under ISO 15189 audit (P<0.001). It can be concluded therefore, that technical standards compliance needs additional attention across the NSW B and G laboratories represented in this study.
Median GGT (%) bias (Figure 7) was consistently between 0.0 and −0.10 suggesting that in general, laboratory measurement of GGT in RCPAQAP samples was under the target value. For B category laboratories, the pattern was similar [bias (%), 0.0 to −0.10], but for G laboratories the medians were between −0.10 and −0.20 showing an increased and significant negative bias compared to the B laboratory time series (Kruskal-Wallis; χ2=61.5, df=1, P<0.001). For serum creatinine, the median (%) bias for all laboratories (Figure 8A) clustered close to 0.0, with the exception of time point 14. For the combined group of 21 laboratories, creatinine concentrations in RCPAQAP samples were close to the target value over repeated testing. When separating into B and G laboratories (Figure 8B), differences were apparent with the general observation that median (%) bias trended above 0.0% for B laboratories, while G laboratory medians were less than 0.0% (again, as exemplified by time point 14). Comparing B and G laboratories across the 16 time points showed a significant difference (Kruskal-Wallis; χ2=74, df=1, P<0.001).
Percentage (%) bias across time for the 2015 RCPAQAP for serum K+ showed less consistency in general, as well as for the B and G laboratory comparison [although at smaller variance compared to GGT and creatinine, ranging from −0.03 to 0.09 (%) bias] (Figure 9). The B category laboratories had seven time points out of 16 where the median was 0.00, with a zero interquartile range (although outliers were present). Taking the number of 0.00 (%) Bias, while B laboratories had seven time points, G laboratories recorded only three time points (3, 14 and 17) at a (%) Bias of 0.00. Apart from the RCPAQAP time points at 0.00, other results for serum K+ overall, and for B/G laboratory categories, were not consistent, showing median and interquartile patterns above and below the (%) bias of 0.00.
To further explore the relationships between ISO 15189 audit and RCPAQAP results, ANCOVA was conducted to explore interactions between GGT (%) bias and other ISO 15189 audit and RCPAQAP variables.
Table 1 summarises the GGT (%) bias model by ANCOVA, where B or G laboratory classification was added as a fixed factor, and bicarbonate (%) bias, K+ (%) bias and total counts of minor (M) reports for laboratories were added as covariates. The ANCOVA model had a Levene’s test significance of 0.321 (F=1.26, df1=3, df2=17), indicating equal variance across the variables. The adjusted R2 was 0.854, and hence explains 85.4% of GGT (%) bias variation among the 21 pathology laboratories evaluated by NATA and EQA (all 21 laboratories were included in the ANOVA, with the influence of B or G lab category assessed as a fixed factor).
Two covariates were significant at P<0.02 [serum K+ (%) bias, and total count of minor reports recorded on ISO 15189 audit inspection], with both variables also recording large effect size (Eta2) results emphasising their strong influence on GGT (%) bias. Bicarbonate (%) bias was added to the model as a control anion, which was not significant; in fact, the removal of bicarbonate bias from the model, or replacement with chloride (%) bias, had negligible impact on the results. Interestingly, the replacement of serum K+ bias with serum sodium (Na+) or calcium (Ca++) bias resulted in a poorer model (adjusted R2<0.70), with neither cation significantly influencing GGT. The replacement of total ISO 15189 audit M count (Table 1) with total ISO 15189 audit C count maintained the quality of the ANOVA results, with a small reduction in adjusted R2. The inclusion of the combined total M and C counts in place of total M or total C alone further reduced the adjusted R2. Further investigation of the impact of measuring M or C ISO 15189 audit results, in relation to EQA performance, requires analysis with a larger data set.
Analyses of ISO 15189 audit reporting focused on minor (M) observations and conditions (C) as markers of NSW laboratory performance, and found that technical ISO 15189 audit criteria recorded significantly more M and C observations compared to laboratory management ISO 15189 audit criteria. EQA data analysed were the total (%) bias measure calculated for all laboratories, for each assay (Figure 6), as well as (%) bias across time for 16 time points representing GGT, serum K+ and serum creatinine, over 2015 for the 21 laboratories included in this study ((Figures 7,8). Interesting temporal patterns were detected for these three assays, with serum K+ proving of particular utility as a sentinel marker of laboratory quality, a position supported by other studies (15,16). ANCOVA showed that serum K+ and the total count (management and technical) of minor (M) observations were significant in a model that explained 85.4% of (%) bias for GGT, demonstrating the close inter-relationship between these three variables measured under ISO 15189 AUDIT and EQA schemes.
Discussion
The quality of pathology laboratory performance is assessed via two processes in Australia, one conducted by NATA that provides written qualitative advice and warnings after a physical inspection of the laboratories, and their management and technical processes. Additionally, the EQA (QAP) process assesses laboratory performance by sending samples of unknown concentration for measurement by individual laboratories. Both processes provide an excellent insight into the quality of pathology laboratories, thus ensuring that the diagnostic information provided to health professionals is of the highest accuracy in terms of patient care. With this foundation, an analytical system that integrates both sources of quality data suggest the opportunity to further enhance the efficacy of quality oversight.
From surveying the literature, there were many reports on the value of high quality results, and in response to this, several proposals on systems to improve quality control. The articles varied from statistical analysis and the proposal of new quantitative models, e.g., Q-Tracks (17), to written reviews or case studies of challenges and responses (15,16,18-20), and comprehensive regional or national level quality assurance programs (21-23). Consistent primary data were not always reported, hence the stratification of the articles into various sub-groups prior to further investigation (Figure 1). The broad nature of these articles as descriptive reports required text analysis to search for themes relating to pathology quality. Common themes were ultimately difficult to find, indicating that in spite of efforts to introduce international harmonisation (e.g., ISO 15189), there are few agreed consistent standards internationally that unify this discipline, as represented by this literature sample.
In this context, there was no strong consensus on how best to proceed, apart from the articles highlighting the value of RCA, as a recommendation to proactively improve quality, or as an effective means to detect specific errors, among other suggestions (11-14). The field, as represented by the 103 articles investigated by text mining, did not provide a consensus on the measurement and analysis of laboratory quality. The high frequency of the words/terms “linearized” and “predictor” clearly demonstrates a desire by clinical scientists to quantitatively define measures of quality in a robust manner.
The statistical models presented here identify markers of poor quality to apply prospectively. Access to predictive rules of quality, emanating from two authoritative sources of evaluation data (ISO 15189 audit and RCPAQAP), allows detection of performance issues well in advance of system failure within a laboratory that will impact on patient care. This information has general applicability for ISO 15189 audit, QAP providers and NPAAC to identify areas where additional monitoring or development of standards may be required. Furthermore, this project will complement NPAAC data gathering used to assess overall laboratory performance in Australia. It will also assist in the development of new standards as areas of poor performance are efficiently and rapidly identified. These metrics should also assist NATA with its audit process.
Machine learning offers a robust statistical base for the detection of patterns and subsequent prediction of outcomes driven by training and testing modalities within the algorithms. Due to the small sample size, only RFA was effective, successfully producing insights into how ISO 15189 audit-generated observations pertaining to laboratory standards link to the quantitative assessment of quality via the EQA. The variation in (%) bias for GGT, serum K+ and serum creatinine were strongly linked to the frequency of minor (M) observations by NATA inspectors, and this RFA showed also that this was vastly superior to an identical model predicting the number of ISO 15189 audit-reported conditions (C). When examining the ISO 15189 clauses it was found that the most frequent problems occurred as non-compliance for technical requirements, in comparison to management, a situation seen also for laboratories in Hong Kong (13). For this study, technical non-conformances were statistically more frequent in comparison the management conditions.
The OOB error rate calculated for RF showed an impressive accuracy for the predictions of low versus high minor (M) condition categories by RCPAQAP markers, entered into the models as a percentage bias values. The same RF investigation, but with condition (C) categories as the dependent variable, were considerably poorer in terms of category prediction accuracy. The RF modelling, therefore, suggests that monitoring of minor (M) conditions is of most value to an integrated predictive model that includes EQA results, expressed as a percentage bias to capture laboratory variation via serum or blood marker detection accuracy. The ANCOVA models that followed used M counts as a response or outcome to explain the significance of ISO 15189 audit measurements in relation to EQA results, with B or G lab classification applied as a main effect or predictor.
While sample size was too small to run SVM (24) and decision trees (8), a successful machine learning proof-of-concept analysis was achieved via RF bootstrapping, which identified GGT, K+, creatinine (%) bias, and ISO 15189 audit M (minor) observations as the leading markers of an integrated ISO 15189 audit and RCPAQAP model. The RFA was successful because the algorithm allows the re-sampling of the two classes over the thousands of decision trees used to calculate the rank of predictors, as well as the prediction accuracy. The suggestion that M observations were the potentially more powerful ISO 15189 audit predictor in this model derived from the low error rate of M class/category prediction (high number of M observations versus low M observation—with a low number of M reports reflecting higher lab quality), compared to the identical model for the prediction of condition (C) categories that had a 43% error rate, and thus poor accuracy. GGT and serum K+ were the leading predictors of M category (Figure 5B). Therefore, a preliminary integrated model of RCPAQAP and ISO 15189 audit data was achieved, which will benefit from a comprehensive future investigation of a full data set from NSW and elsewhere.
The subsequent ANCOVA models revealed that to explain GGT % bias, the number of minor (M) conditions recorded, expressed as a count and not a category, was a significant predictor of GGT bias. Interestingly, from the other RCPAQAP % bias results, serum K+ was highly significant (P<0.001), suggesting a strong relationship between GGT and K+ for the EQA quality assessment process. Laboratory type (B or G) was not significant. The strength of relationship was suggested also by the adjusted R2 for the total model (0.854), which was not retained, for example, if Na+ was substituted for K+, or C conditions instead of M conditions.
The value of serum K+ has been previously recognised by Meier et al. (17), regarding effective index markers of quality, and suggesting that serum K+ may be the ideal longitudinal EQA marker. Achieving a zero percent bias (0.00%) is possible, as demonstrated particularly by B laboratory results (Figure 9B). When K+ bias variation occurs, it appears to arise from laboratory results that both over- and under-estimate the target value set by the EQA. Variation due to bias and precision have been explored and commented upon by other studies, and emphasise the value of serum K+ as a useful quality marker (25,26).
This paper has quantified laboratory quality through the use of (%) bias and counts of C and M under ISO15189. The desire to introduce robust quantitative measures to assess lab quality has recently also produced the failure mode and effects analysis (FMEA), which calculates a risk based on a hierarchy of untoward process events (incidents), and the event types’ different contributions to total risk (27,28). Combinations of monitored incident frequencies, estimated detection difficulties, and rank ordering harms of untoward events, integrate incident monitoring and risk calculation. In human factors engineering, FMEA calculates risks to each error-type under study, by multiplying objective error event-probabilities (cause-occurrence frequencies) by subjective harm-probabilities and subjective detection difficulty scores (both on a 1–10 scale). The first step is to measure the objective probability of failure (error-event frequency) at each step in a process implicated by detected error. The second step, guided by medical judgment, requires an estimate of how likely specific failure(s) in an event-sequence are produced, resulting in harm-outcome(s) and, also, how difficult it is to discover this connection, where after the second medical judgment assigns a detection difficulty to discovering the connection between the process error and harm. The third FMEA step assigns levels of severity to the defect(s) (harm-outcome weights).
In conclusion, this pilot study has systematically appraised the international pathology quality literature up until late 2016, and found that while there were many excellent insights into questions of performance, there was no clear consensus on how to optimise quality assurance across the various inspection, monitoring and measurement procedures. In response, we have developed a preliminary machine learning (RFA) centric model with which to assess the relationship between ISO 15189 audit observations and EQA performance, as represented by (%) bias found for assays requested by the Australian EQA process (via the RCPAQAP). The results indicated that the number of minor ISO 15189 audit observations were the best measure to link to EQA results, and for this study, GGT, serum creatinine and serum K+ were the best predictors of M, with an overall accuracy of almost 85% (with poor prediction accuracy for C category at around 57%). Others have suggested that serum K+ is an ideal sentinel marker of pathology quality (25). The interrelationships between ISO 15189 audit M category, GGT and K+ were supported also by the results of ANCOVA, which resulted in a robust model with an adjusted R2 of 85%. Future studies on data from larger laboratory cohorts are required to validate these results, as well as repeat modelling using % bias data calculated from specific EQA target values, provided by the RCPAQAP.
Acknowledgments
Funding: Many thanks to the Quality Use of Pathology Program (QUPP), the Commonwealth Department of Health (Australia), who awarded funding to support this project (No. 4-2UJWED1).
Thanks also to NSW Health Pathology and the Royal College of Pathologists of Australasia Quality Assurance Programs (RCPAQAP) for access to anonymous NATA and EQA data. The staff in the ANU (JCSMR and RSPH) Research, Human Resources and Finance Offices, thank you for general administrative support and the facilitation of contracts, payments and agreements between the ANU and the Commonwealth Department of Health. Also, the authors wish to acknowledge the contribution of Guifang Shang in the early preparation of the article library used for text mining.
Footnote
Conflicts of Interest: Tony Badrick serves as an unpaid editorial board member of Journal of Laboratory and Precision Medicine from December 2016 to November 2018. T Badrick is the CEO of the Royal College of Pathologists of Australasia (RCPA) quality assurance program (QAP), G Koerbin is the Chief Scientist of NSW Health Pathology. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- NATA. National Association of Testing Authorities, Australia. 2017. Available online: https://www.nata.com.au/nata/
- Feinerer I, Hornik K. tm: Text Mining Package. R package version 0.6-2. 2015. Available online: https://CRAN.R-project.org/package=tm
- Feinerer I, Hornik K, Meyer D. Text Mining Infrastructure in R. J Stat Softw 2008;25:1-54. [Crossref]
- Bouchet-Valat M. SnowballC: Snowball stemmers based on the C libstemmer UTF-8 library. 2014. Available online: https://CRAN.R-project.org/package=SnowballC. Accessed 05/09/2017
- Wickham H. ggplot2: Elegant Graphics for Data Analysis Springer-Verlag. New York: Springer-Verlag, 2009.
- Fellows I. Wordcloud: Word Clouds. R package version 2.5. 2014. Available online: https://CRAN.R-project.org/package=wordcloud
- Maechler M, Rousseeuw P, Struyf A, et al. cluster: "Finding Groups in Data": Cluster Analysis Extended Rousseeuw et al. R package version 2.0.6. 2017. Available online: https://cran.r-project.org/web/packages/cluster/index.html
- Breiman L Friedman JH, Olshen RA, et al. Classification and Regression Trees. Belmont, CA: The Wadsworth and Brook, 1984.
- Liaw A, Wiener M. Classification and regression by randomForest. R news 2002;2:18-22.
- IBM SPSS Statistics for Macintosh, Version 22.0. Armonk, NY: IBM Corp, 2013.
- Bhat V, Chavan P, Naresh C, et al. The External Quality Assessment Scheme (EQAS): Experiences of a medium sized accredited laboratory. Clin Chim Acta 2015;446:61-3. [Crossref] [PubMed]
- Allen LC. Role of a quality management system in improving patient safety - laboratory aspects. Clin Biochem 2013;46:1187-93. [Crossref] [PubMed]
- Ho B, Ho E. The most common nonconformities encountered during the assessments of medical laboratories in Hong Kong using ISO 15189 as accreditation criteria. Biochem Med (Zagreb) 2012;22:247-57. [Crossref] [PubMed]
- White B. The impact of ISO 15189 and ISO 9001 quality management systems on reducing errors. Vox Sang 2002;83:17-20. [Crossref] [PubMed]
- Ahmad M, Khan FA, Ahmad SA. Standardization of pathology laboratories in Pakistan: problems and prospects. Clin Biochem 2009;42:259-62. [Crossref] [PubMed]
- Unsal I, Fraterman A, Kayihan I, et al. ISO 15189 accreditation in medical laboratories: An institutional experience from Turkey. Clin Biochem 2009;42:304-5. [Crossref] [PubMed]
- Meier FA, Souers RJ, Howanitz PJ, et al. Seven Q-Tracks monitors of laboratory quality drive general performance improvement: experience from the College of American Pathologists Q-Tracks program 1999-2011. Arch Pathol Lab Med 2015;139:762-75. [Crossref] [PubMed]
- Albertini A, Signorini C. The quality assurance system in clinical chemistry. Ann Ist Super Sanita 1995;31:3-8. [PubMed]
- Sierra-Amor RI. Mexican experience on laboratory accreditation according to ISO 15189:2003. Clin Biochem 2009;42:318. [Crossref] [PubMed]
- Plebani M, Sciacovelli L, Chiozza ML, et al. Once upon a time: a tale of ISO 15189 accreditation. Clin Chem Lab Med 2015;53:1127-9. [Crossref] [PubMed]
- Morisi G, Leonetti G, Palombella D, et al. Organization and results of a pilot scheme for external quality assessment in clinical chemistry carried out in the Latium region, Italy. Ann Ist Super Sanita 1995;31:113-22. [PubMed]
- Baadenhuijsen H, Kuypers A, Weykamp C, et al. External Quality Assessment in The Netherlands: time to introduce commutable survey specimens. Lessons from the Dutch "Calibration 2000" project. Clin Chem Lab Med 2005;43:304-7. [Crossref] [PubMed]
- Haeckel R, Wosniok W, Gurr E, et al. Permissible limits for uncertainty of measurement in laboratory medicine. Clin Chem Lab Med 2015;53:1161-71. [Crossref] [PubMed]
- Karatzoglou A, Meyer D, Hornik K. Support Vector Machines in R. J Stat Softw 2006;15:1-28. [Crossref]
- Haag MD, Kelly JR, Ho A, et al. A study to examine the accuracy of potassium measurements in clinical laboratories across Canada. Clin Biochem. 2000;33:449-56. [Crossref] [PubMed]
- Kost GJ, Hale KN. Global trends in critical values practices and their harmonization. Clin Chem Lab Med 2011;49:167-76. [Crossref] [PubMed]
- Mackay M, Hegedus G, Badrick T. A simple matrix of analytical performance to identify assays that risk patients using External Quality Assurance Program data. Clin Biochem 2016;49:596-600. [Crossref] [PubMed]
- Badrick T, Gay S, Mackay M, et al. The key incident monitoring and management system - history and role in quality improvement. Clin Chem Lab Med 2017; [Epub ahead of print]. [Crossref] [PubMed]
Cite this article as: Lidbury BA, Koerbin G, Richardson AM, Badrick T. Integration of ISO 15189 and external quality assurance data to assist the detection of poor laboratory performance in NSW, Australia. J Lab Precis Med 2017;2:97.


