
DNA damage assessment and potential applications in laboratory diagnostics and precision medicine
Establishment of new biomarkers in precision medicine
Personalized medicine, often referred to as precision medicine in the Anglo-Saxon literature, represents a new paradigm in medicine. In contrast to the “one-fits-all” approach, precision medicine takes individual variability of patients into account. It focuses on the classification of patients with the same disease into subgroups after determination of specific diagnostic, predictive or prognostic biomarkers (1). Depending on these individual patient profiles, precision medicine finally tries to provide tailored treatment strategies (2).
Despite the common use of the terminology, there is still an ongoing debate on the exact definition of this new strategy (3). Attempts to narrow the scope of precision medicine to genetics only fall definitely too short. Precision medicine should be considered as a medical model using cellular characteristics and molecular profiling for tailoring the right therapeutic strategy for each individual person at the right time, and/or for determining the predisposition to disease, and/or for delivering early and targeted prevention (1).
Emerging progress in “- omics” technologies has paved the way for a more comprehensive biomarker-based stratification of patients. Novel diagnostic techniques have entered the limelight offering the analysis of new biomarkers and thereby demanding a new awareness in laboratory medicine and beyond.
In this respect, one of the most intriguing developments has taken place in the field of precision cancer therapy. Although tumor heterogeneity represents a major obstacle for precision medicine, much progress has been achieved in treating cancer patients based on individual molecular profiles (4). This development has been accompanied by the search for new biomarkers. The prediction of individual responses to anticancer therapy has been in the focus of research highlighting the need for a profound understanding of drug response mechanisms to identify new biomarkers (5).
Whereas pharmacogenetic analyses of certain mutations are already included as biomarkers in so-called companion diagnostics, functional tests assessing the level of induced DNA damage and DNA repair capacity might potentially be used in clinics as well.
According to guidelines for “in vitro companion diagnostic devices” published by the U.S. Food and Drug Administration (FDA) such diagnostic tests should for instance identify populations who are (I) either likely to benefit; (II) be at increased risk for serious adverse effects for a particular treatment; or (III) biomarker assessment should improve safety or efficacy by monitoring treatment response (6). Furthermore, apart from proven clinical value, Taube et al. (7) stated two additional criteria in that context. Robustness and reproducibility of an assay accompanied by the conviction and acceptance of this test among the clinical community are prerequisites for a successful translation. As a fact, based on past experience, difficulties would be expected to achieve standardization of genotoxicity assays in the near future, not least because scientists would be reluctant to change their methods (8).
Recently, the Organization for Economic Co-operation and Development (OECD) updated or newly published guidelines about chemical testing for multiple genotoxicity assays. Guidance on assay performance may help to standardize these tests to gain more information about their reliability as diagnostic parameter and for translation into clinical laboratories. Thereby, biomarkers of DNA damage response (DDR) can potentially help to predict individual sensitivity or resistance of normal tissue and tumor cells regarding chemo- or radiotherapeutic anti-cancer treatment. Further, DDR markers can support monitoring of treatment efficacy and to adjusting treatment schedules (9).
DNA damage and DNA damage response
The DNA of every cell is continuously damaged by endogenous sources, such as replication or by metabolic (by-) products [(e.g., reactive oxygen species (ROS)]. Additionally, multiple exogenous factors, like ultraviolet light, ionizing radiation, various genotoxic drugs and environmental toxins are capable to induce DNA lesions (10). In contrast to other biomolecules which will be degraded and newly synthesized after alteration, DNA does not underlie such constant recycling process. Instead, a variety of lesion-specific DDR mechanisms exists to restore DNA integrity. During the last decades, innumerous studies uncovered diverse molecular DDR mechanisms which have been extensively reviewed previously (10,11). One of the most severe types of DNA damage are DNA double-strand breaks (DSBs). They are either repaired by classical or alternative non-homologous end-joining (NHEJ) or by homologous recombination (HR) (12). Nonetheless, DNA damage accumulates throughout lifetime and induces chromatin alterations in different cell types, such as tissue-specific stem cells. This may be the driving force of aging as well as for the development of numerous diseases, like malignancies (13,14). Defects within DDR pathways have been reported to be involved in tumorigenesis and premature aging (15,16).
In contrast to normal tissue where DNA damage should be avoided, certain therapeutic strategies are based on DNA damage induction in pathologic cells. Thus, anti-cancer chemo- or radiotherapies often take advantage of DDR defects of tumor cells to specifically target and kill malignant cells (17). The tremendous progress in the understanding of DNA damage signaling has led to new therapeutic options in particular for cancer treatment by modulating these DNA repair pathways, such as the therapeutic concept of synthetic lethality (11,17).
Genotoxicity assays
Treatment with exogenous DNA damage-inducing agents like cytostatic drugs or ionizing radiation can be toxic for cells and represents the basis for anti-cancer therapy. Genotoxicity tests analyze transient or permanent defects of the genetic material (18). However, genotoxicity does not need to be accompanied by mutagenesis or cytotoxicity nor does cytotoxicity need to be caused by genotoxic effects.
Besides genotoxicity reporter assays employed in bacteria (e.g., Ames test) or cell lines (e.g., mouse lymphoma thymidine kinase assay), several techniques can be applied to analyze DNA damage and corresponding DDR in primary human cells. The frequency of cytogenetically detectable irreversible chromosomal damage can be assessed for instance by chromosomal aberration (CA), especially by dicentric chromosome assay (DCA) (Figure 1A) or by cytokinesis-block micronucleus assay (CBMN) (Figure 1B). Further, molecular genotoxicity assays can be used to study various endpoints. They can be employed either as indicators for primary DNA damage before occurrence of DNA repair or to analyze remaining DNA lesions several days after treatment. Two of the most commonly used tests are the single-cell gel electrophoresis/comet assay (COM) (Figure 1C) and γH2AX immunofluorescence microscopy analysis (Figure 1D). Each method has its own characteristic assay performance regarding specificity and sensitivity. Consequently, depending on the study and targeted endpoint, an appropriate assay should be selected. Concerning their potential use for precision medicine, these four methods will be described in greater detail below. In fact, various other tests were applied in studies to examine genotoxic effects, such as sister chromatin exchange, the Halo assay or the application of electrochemical methods, polymerase chain reaction, high performance liquid chromatography or mass spectrometry (19,20). However, these assay techniques are beyond the scope of this review.
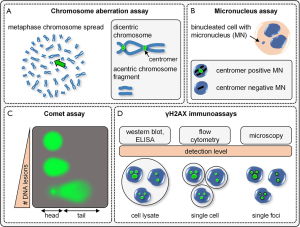
Chromosome aberration assay
The CA assay has been applied for chemical testing for more than 40 years and still remains the gold standard to assess genotoxicity particularly for radiation biodosimetry (21,22). Apart from the analysis of different numerical chromosome and chromatid variants as well as other abnormalities (e.g., DNA strand breaks or translocations), the quantification of chromosome type changes [e.g., rings and mainly dicentric chromosomes (DIC)] remains the method of choice (Figure 1A) (21,23). The dicentric chromosome assay (DCA) is based on misrepair and fusion of DNA double-stranded ends of two different chromosomes, resulting in the formation of dicentric (2 centromeres) chromosomes and accompanying acentric chromosomal fragments. This analysis is widely performed on peripheral blood lymphocytes which are stimulated for 48 h and arrested in metaphase. Microscopic evaluation of the metaphase chromosome spread is commonly performed after a Giemsa (G)-band staining (21,24). To improve the sensitivity and allow the detection of chromosomal translocations or deletions, CA testing can be combined with specific chromosomal staining by fluorescence in situ hybridization (FISH) (25). Thus, FISH using centromere or telomere-specific probes are widely applied for DCA. Although CA testing is characterized by a high sensitivity, it is subjected to pre-analytical variation due to the required cell culture. Further, the microscopic chromosome spread analysis is influenced by operator subjectivity and potential bias, depending on the level of observer experience (26). Finally, the late endpoint examined by CA assessment does allow neither initial DNA damage analysis nor repair kinetic studies.
Cytokinesis-block micronucleus assay
CBMN is another widely accepted cytogenetic method to study chromosomal damage or loss, mitotic dysfunction and cell death (Figure 1B) (27). CBMN can be performed with various human cell types whereas the use of lymphocytes, either isolated or within whole blood cultures, remains the preferred approach (24). Thus, lymphocytes are stimulated for 72 h and treated after 44 h with a cytokinesis inhibitor (e.g., cytochalasin B) to block the division into two daughter cells. Cells that underwent the first division of the nucleus can be identified as binucleated cells. These cells are screened for micronuclei (MN). A MN represents a whole chromosome or chromosomal fragment engulfed in a small extranuclear body that was separated from the nucleus during mitosis (Figure 1B).
Furthermore, aneugenic and clastogenic effects can be differentiated by additional centromere assessment using immunofluorescence kinetochor staining or FISH technology (28). Whereas aneugenic mechanisms interfere with the mitotic apparatus, which leads to the loss of whole chromosomes and centromere-positive MN, clastogenic effects cause chromosomal disruptions and breakage resulting in centromere-negative MN (29,30). Apart from centromeric labeling, the FISH technique can be used for whole chromosome staining. Thus, further information on chromosomes or fragments encapsulated in the MN can be obtained. To assess the mutagenic potential in in vitro pharmaceutical testing, reagents under investigation often require additional exogenous metabolic activation to induce genotoxicity. Therefore, exogenous enzymes often obtained from the S9 fraction of rodent liver homogenate are used in many pharmacological studies to mimic human liver metabolism (31).
MN formation can be caused by different lesions of both the spindle apparatus and the DNA. Although chromosomal breakage in general results from DNA DSBs, these can also originate from base lesions or single-strand breaks (SSBs) subsequently converted into DSBs during replication, especially when DDR is impaired. Compared to CA testing, evaluation of the MN assay is less technically demanding. The major shortcomings of MN detection, however, are the late endpoint analysis after 3 days and the lack of information on initial DNA damage before the beginning of DDR (32). Further, for an adequate MN evaluation, a minimum of 1,000 binucleated cells should be analyzed.
Besides scoring of MN, more information can be retrieved by analyzing additional structures like the formation of nucleoplasmic bridges (NPBs) or nuclear bodies (NUBDs) indicating gene amplification or DNA disrepair, respectively. Further, quantification of mono-, bi- and multinucleated cell ratio enables proliferation studies by calculation of the nuclear division index (NDI) (27).
Single-cell gel electrophoresis (comet assay)
A whole array of different DNA lesions like SSB and DSB, alkali-labile DNA sites as well as uncompleted base excision repair (BER) can be detected by alkaline (pH >13) single-cell gel electrophoresis (33). Apart from the classical pulsed-field gel electrophoresis (PFGE) technique, where DNA fragments of a pooled cell extract are separated by an alternating electric field within an agarose gel, single cell gel electrophoresis, also referred to as comet assay, enables the analysis of DNA lesions in individual cells. As a fact, this assay technique is a widely accepted genotoxicity method. The comet assay can be performed with various cell types and does not require cell proliferation. Cells are embedded in low melting agarose on a slide and the cell membrane, cytoplasm, and nucleoplasm are removed by lysis with hypertonic, non-ionic detergents. After alkaline treatment, the unwounded, denatured negatively charged DNA remains in the center of the cell and starts to migrate toward the anode during electrophoresis (34). The smaller the size of the DNA fragments the faster the movement. This leads to the formation of comet-like tails in damaged cells, which can be stained with DNA dyes and quantified via microscopy and image analysis (Figure 1C).
Several methodological modifications have been described for the comet assay. It has been claimed that the comet assay under neutral conditions preferentially leads to the detection of DSBs. However, this does not seem to hold true since SSBs as well as DSBs cannot be distinguished either in alkaline nor in neutral comet assays (34). Of note, both assay conditions for single-cell gel electrophoresis are characterized by different assay performances resulting in different comet shapes and sensitivities (33). To increase the spectrum of DNA lesions measurable with the comet assay, protocols have been established applying DNA digestion with different lesion-specific endonucleases which enable conversion of oxidized bases into detectable SSBs (35). Furthermore, a combination of the comet assay with FISH was described to study DDR in particular genes or DNA sequences (36).
In general, comets of 100 cells per sample need to be analyzed. Depending on the evaluation method different parameters can be determined. In a semi-quantitative manner, comet images can be classified typically into five distinct categories according to their relative tail intensity. Moreover, modern image analysis enables the measurement of tail length, total intensity, percentage of DNA localized in the head and tail as well as the product of tail length and DNA content, referred to as olive tail moment (34).
Altogether, the comet assay is a sensitive and user-friendly assay for detecting DNA damage and can be performed readily with moderate costs (24). In contrast to CA and MN assays, the level of initial DNA damage can be analyzed by comet assay directly after treatment before main initiation of DNA repair. Of note, different studies revealed a high inter- and intra-laboratory variation. For better reproducibility, assay conditions need to be further standardized (37). Additionally, cell death needs to be controlled since apoptotic cells can lead to false-positive findings (26).
γH2AX immunocytochemistry assay
The latest genotoxicity assay described here, is based on the immunocytochemical detection of the phosphorylated histone variant γH2AX (38). Upon DSB formation, H2AX molecules in the chromatin surrounding the DSB site become rapidly phosphorylated at serine 139 by the PI3-like kinases, ataxia telangiectasia mutated (ATM), ATM- and Rad3-related kinase (ATR), or DNA-dependent protein kinase (DNA-PK). A feedback loop leads to signal amplification and the formation of γH2AX foci which can be visualized as discrete spots after specific immunofluorescence staining (39). Unlike cytogenetic assays, where genotoxicity is assessed at an endpoint distant from the initial DNA damage, the γH2AX assay can be applied at different time points, either to study primary damage, repair kinetics or residual levels of DSBs (40,41). Furthermore, focus formation does not require cell cycling and can be observed in non-proliferating as well as in proliferating cells.
However, γH2AX analysis is limited to the detection of DSBs and there is evidence that formation of foci can also occur in the absence of DNA damage (42). Recent research indicates that many proteins interacting with DNA damage-modified histones do not directly participate in DNA repair. Instead, spreading of chromatin modifications away from the primary lesions may be an auxiliary mechanism evolved to coordinate repair with transcription and replication, as reported by Nakamura et al. (43) and Polo et al. (44).
Different immunological methods can be employed to determine the level of γH2AX. Whereas enzyme-linked immunosorbent assay (ELISA) or immunoblotting enable the analysis of pooled cell or tissue extracts only, individual cells can be examined by flow cytometry or fluorescence microscopy (Figure 1D). Analysis of γH2AX by flow cytometry allows a fast, high-throughput testing assessing thousands of events per sample. It can be combined readily with cell-cycle studies by measuring DNA content or classification of different subpopulations, e.g., by applying specific cell surface markers. Flow cytometry is the method of choice for doses beyond 2 Gy, where classical γH2AX foci quantification via microscopy assessment becomes imprecise (45). In contrast, fluorescence microscopy is reported to be the most sensitive method, enabling the detection of down to only a single focus per cell (46). Therefore, it is especially applicable for the investigation of cells expressing low numbers of γH2AX foci. Other advantages of fluorescent microscopy analyses are the feasibility of co-localization studies with additional DDR proteins as well as examination of morphology and spatial distribution of individual foci. Of note, acceptable intra- and inter-assay variability for the analysis of γH2AX in peripheral blood lymphocytes after ionizing radiation has been reported in various studies (47,48). In addition, this approach offers a substantial advantage regarding the statistical analysis of data. Commonly obtained parameters are central tendency (e.g., mean or median) and variation (standard deviation) of the quantified amount of foci (49,50). Instead of using relative intensity values or qualitative descriptions, γH2AX foci analyses enable quantification of absolute numbers (number of foci per cell). These quantitative data allow statistical calculations based on Poisson distribution and kinetical analysis by non-linear regression. Thus, the probability of a given number of foci occurring in a fixed interval of cells can be calculated and even significantly over-dispersed foci/intensity distributions can be accurately assessed (49,50). These data can be readily combined with other (co-localized) parameters [e.g., p53 binding protein 1 (53BP1)] which enables a high level of accuracy (51).
It was stated that γH2AX foci quantification using fluorescence microscopy represents the most sensitive approach, since single foci can be detected (46,52). But direct comparative analyses of more than two techniques for γH2AX detection regarding assay performance have been lacking so far. Thus, the technical report by Reddig et al. in this issue of the journal provides the first comprehensive comparison in terms of sensitivity of γH2AX detection by immunoblotting, flow cytometry and immunofluorescence staining with automated foci quantification and fluorescence detection of etoposide-exposed lymphocytes.
Of note, recent electron microscopic studies have provided a more detailed picture of the spatial arrangements of repair proteins within DNA damage foci. Electron microscopy is a technically demanding and challenging technique and, thus, its use for precision medicine will depend on its cost-effectiveness and availability. Nevertheless, the high resolution of transmission electron microscopy (TEM) permits the visualization of gold-labelled repair proteins at the single molecule level and to characterize the spatiotemporal dynamics of DSB induction and repair within the chromatin ultrastructure (53,54). DNA damage signaling and DNA repair dynamics differ significantly in DSBs located in different chromatin environments such as hetero- and euchromatin. Even highly clustered DNA lesions induced by densely ionizing radiation that appeared as a single focus by fluorescent microscopy can be dissolved as multiple DSBs in close proximity by this TEM approach (55). The high resolution of TEM permits the visualization of the essential components of the DNA repair machinery at the single molecule level, such as the Ku70/Ku80 heterodimer for classical NHEJ (53,54). By labeling the activated Ku heterodimer, which binds directly to broken DNA ends in preparation for rejoining, this TEM approach permits the reliable detection of actual DSBs (14,55). Further, in proof-of-concept studies, the γH2AX assay was employed on six cancer cell lines to investigate off-target effects by genome editing with CRISPR/Cas9 (56,57). Altogether, despite some limitations, the γH2AX assay is so far the most sensitive and specific test for detecting DNA DSBs (42,58).
Standardization and automation of genotoxicity assays
Regarding automation and standardization requirements, one limitation CA, CBMN, comet assay and γH2AX immunofluorescence tests have in common is their labor-intensive, manual and subjective microscopic analysis. To enable high-throughput screening and standardized assay interpretation, much effort has been made to develop automated approaches. Whereas some software programs only enable semi- or fully automated evaluation of prior separately acquired microscopy images, other tools combine image acquisition, processing and analysis.
In general, automated analysis requires fixed cell samples immobilized on microscopy slides at the particular endpoint according to the corresponding protocol of the applied method. Except for Giemsa stain, staining is predominantly conducted with fluorescence DNA dyes, like 4',6-diamidino-2-phenylindole (DAPI), acridine orange, SYBR green/gold or fluorescence-labeled probes and antibodies, which later mark the regions of interest. For automated evaluation, stained samples are processed by an interpretation system comprising mainly a motorized fluorescence microscope combined with an appropriate camera and equipment for image acquisition. All devices are connected to a computer to provide sufficient data storage and software modules to control hardware components, automated image acquisition, analysis, and evaluation. Essential for reliable analysis are image quality and the accuracy of implemented algorithms required for image segmentation and pattern recognition. Thereby, suitable objects, such as metaphase chromosomes, bi-nucleated cells, comets or cell nuclei as well as characteristic regions representing DNA lesions, like DIC, MN, comet tails or foci will be identified and evaluated.
In the past, various instruments and software tools have been developed for automated genotoxicity assessment. Some focused on the fast and standardized identification of DIC in classical metaphase spreads (59,60) or directly in interphase cells, applying premature chromosome condensation (PCC), a technique to allow immediate damage detection without additional lymphocyte stimulation (61). Multiple tools have also been developed for automated micronucleus analysis, either based on fluorescence microscopy (62,63) or imaging flow cytometry (64). In a report published 2013 by Fenech et al., the authors compared different systems available for MN scoring (65).
In conventional comet assays, only one or two samples can be placed on one microscopy slide, only a small fraction of cells is analyzed and the space in electrophoresis tank is limited. Hence, different approaches using, e.g., 12 minigels per slide or 96-well microplate format (35) and a CometChip assay (66) have been developed to increase the throughput of samples. To avoid random distribution and overlap of cells within the gel, the CometChip utilizes agarose gels with micropores as small as a single cell to generate a consistent cell grid (66). This controlled cell arrangement combined with a 96-microwell setting generated a platform for fast processing of large sample numbers. Further, the CometChip was shown to be a powerful tool to assess genotoxicity mediated by engineered nanoparticles (67). Therefore, single- and double-stranded DNA breaks, alkali-sensitive sites as well as variations in DNA repair pathways are detectable by comet assay at a high-throughput level.
Since manual scoring of comets can only be performed in a semi-quantitative manner multiple commercially available software tools and free-ware options have been designed for automated image analysis enabling comet size and intensity measurements (59,68,69). For further standardization, networks and workshops were established to exchange data and knowledge among different research groups regarding application and technical issues (70,71).
Whereas international guidelines were released, e.g., by the OECD for the genotoxicity assays mentioned above no such guidelines exist for γH2AX foci analysis. Besides differences in assays performance also manual quantification of γH2AX foci may result in high intra- and inter-laboratory variability. Within the last 15 years several tools have been developed to enable automated γH2AX foci quantification. Most applications comprise software programs for digital image processing and analysis which automatically detect nuclei and corresponding foci. Here, images needed to be acquired separately on a fluorescence microscope, as described in detail elsewhere (72-75). In contrast, different fully automated microscope systems were designed or modified combining image acquisition as well as foci analysis without required presence of operator during the scanning process (59,76,77). Furthermore, a Rapid Automated Biodosimetry Tool (RABiT) for large-scale biodosimetry studies was constructed. This robotic workstation implements fully automated sample processing, including lymphocyte isolation from fingerstick-derived blood samples and antibody staining, as well as image acquisition and γH2AX analysis (78).
These approaches may facilitate the standardization of the γH2AX assay and its translation into clinical diagnostics, as shown for different immunofluorescence tests in the field of autoimmune diagnostics. To design a reliable automated analysis platform, criteria for selection or exclusion of objects as well as for object analysis need to be defined conscientiously and must be implemented in adequate automated interpretation systems. But once achieved, these approaches offer standardization, objectivity, and reproducibility as has been demonstrated recently for autoantibody testing by indirect immunofluorescence employing novel automated interpretation platforms, like the AKLIDES system (79,80). Within a short period of time, automated pattern interpretation has been introduced into routine autoimmune testing. Indeed, this development has ushered in a new era in this field of laboratory diagnostics (81). Altogether, a subjective interpretation analysis has been turned into a standardized automated technique. Similarly, the recent progress in the automated interpretation of genotoxic assays like the γH2AX assay has provided promising results. Several studies demonstrated a satisfactory correlation between manual and automated interpretation supporting this assumption (76,82). Thus, these new interpretation technologies seem to be on the brink of translation into precision medicine especially in the field of oncology (83).
Fully automatized high-throughput assays such as the CometChip or the γH2AX assays are a significant contribution to precision medicine since they enable a comprehensive determination of the response upon drug treatment. In particular γH2AX foci assessment can be combined with co-localization analysis of associated biomarkers, such as 53BP1, and provides information on the phenotypic (cell shape, nucleus diameter) and the proteomic level (relative expression level of a biomarker). The comet and γH2AX assays assess the DNA damage of single cells with a high redundancy (>100 cells/test), high replicate number as well as on different cell populations and at multiple time points after treatment. This means that dose-limiting side effects may be quantified for each patient. In particular, the response to chemotherapy or radiation therapy potentially benefits from this information gain (84,85).
Nonetheless, further optimization of standardized criteria for assay performance and evaluation will improve the comparability of automated analyses and pave the way for multi-center studies (65). The novel automated systems available should be able to meet the demands of modern data exchange to facilitate the management of data pooling in the framework of “big data” regarding urgently needed multicenter studies for biomarker evaluation (86). Accordingly, DNA damage assays should be assessed depending on their application to demonstrate their usefulness to provide diagnostic, prognostic, and/or predictive biomarkers for precision medicine (87).
Genotoxicity assays in laboratory diagnostics and precision medicine
Ineffective DNA repair mechanisms and genomic instability are fundamental characteristics of different human diseases. Whereas altered DDR can lead to accumulation of DNA lesions and apoptosis of irreplaceable neurons in neurodegenerative disorders like Alzheimer’s disease, infinite growth of mutated cells represents a typical feature of neoplastic diseases (88). To replace traditional, uniform disease classification and treatment, especially in cancer therapy, a paradigm shift needs to take place. The high heterogeneity among tumors should be addressed by a more comprehensive biomarker-based stratification of patients in the framework of precision medicine. Besides basic research and pharmacological testing, genotoxicity assays, especially γH2AX, offer a wide range of applications in the field of radiation biodosimetry, occupational/environmental exposure to potentially genotoxic agents and clinical studies (89,90). However, apart from CA assays in prenatal diagnostics, these genotoxicity assays have not been widely transferred into clinical routine yet, not least because of insufficient standardization and the lack of data from well-characterized clinical studies.
One potential application of genotoxicity tests is the use as biodosimetry tool for the triage and classification of exposed individuals after a major nuclear or radiological emergency, to rapidly identify and treat highly irradiated subjects. In this regard, projects between multiple European laboratories have been established for validation and harmonization of different genotoxicity assays (91,92). Retrospective dose estimation confirmed highest accuracy for DCA but also reported a feasible application of more rapid γH2AX tests, especially for screening and identification of most severely exposed subjects (22,93). In comparison to DCA, CBMN or COM the γH2AX assay was characterized by its high sensitivity enabling detection of initial DNA damage even after low dose of X-ray exposure (1–10 mG), as it is applied in clinical radiography (94,95).
Besides the absorbed radiation dose also individually varying biological mechanisms affect the degree of radiation damage, resulting in a wide spectrum of acute and late tissue responses. Different functional assays have been developed for prediction of tumor and normal tissue radiosensitivity employing ex vivo irradiation of patient samples. Assessment of CA, MN formation and clonogenic survival represent the most established techniques for prediction of radiosensitivity but endpoint analysis requires several days to weeks (96). Therefore, multiple studies have been conducted investigating the prediction of radiosensitivity also by comet or γH2AX assay (97,98).
One of the most promising approaches of these two assays in clinics is the prediction of normal tissue toxicity and identification of subjects which carry a hetero- or homozygote defect in DNA damage responses genes, e.g., Ataxia telangiectasia (AT). These patients, especially children, are at high risk to develop severe radiation toxicities. A majority of these subjects could be identified in several studies by their reduced DNA damage repair kinetics after ex vivo cell radiation by comet (99,100) and γH2AX assays (53,101). Especially residual γH2AX foci represented a promising biomarker for prediction of normal tissue radiosensitivity, although assessment alone could not be correlated to over-responders in all conducted trials (102,103). Since many intrinsic and microenvironment-dependent factors influence tumor response to radiation therapy genotoxicity assays have also been tested for their ability to predict tumor radiosensitivity. Discrimination of radiosensitive and radioresistant tumor cell lines or tissue samples was successfully demonstrated by comet (97,104) and γH2AX assays (41,103) but further validation and standardization are required for these tests to be transferred into laboratory diagnostics and precision medicine. Lacombe et al. (105) comprehensively reviewed different assays and biomarkers used to predict tumor radiosensitivity in this context.
Another obstacle in cancer treatment are individually varying intrinsic or acquired mechanisms leading to modulated chemoresponsiveness, such as alterations in DDR, apoptosis or expression level of drug efflux transporters (106). Different experiments using either the comet or γH2AX assay have demonstrated the usefulness of these methods to assess drug efficacy of DNA damaging agents as well as modulation of chemoresistance (84,107-109). Further, γH2AX foci analysis by immunofluorescence staining has been reported to provide a promising biomarker to predict the response of patients to ionizing radiation and to analyze the combined effect of radiotherapy and DDR-modifying drugs (45). Thereby, new information on single and combined drug toxicity and on their pharmacokinetics can be obtained. Additionally, analysis of DICs was proposed to be a potential technique for the follow-up of patients after genotoxic drug treatment (61).
Besides applications in the field of oncology, genotoxicity assays also show potential to be used as diagnostic tools of chronic inflammation or other age-associated diseases, such as autoimmunity or neurodegenerative disorders (110-112). Furthermore, assessment of genomic integrity may be used as prognostic indicator of different malignancies developing later in life, which is associated with DNA damage induced by various epidemiological factors, such as chronic inflammation, obesity, smoking or occupational exposure to radiation or toxins (89). Another application of genotoxicity analysis has been reported in sports medicine as a marker of DNA damage in response to aerobic physical exercise (113-115). Further, the high-sensitive detection of γH2AX foci has been used to investigate the safety profile of non-ionizing radiation after high-field and ultra-high-field strength magnetic resonance imaging and mobile phone radiofrequency exposure (116-119).
Currently, precision cancer treatment is mainly focusing on genetic and epigenetic analyses. Even though this strategy seems very promising, identification, verification, and interpretation of candidate DNA sequences as biomarkers are difficult and still do not always reveal the level of protein expression and activity. In contrast, the genotoxicity assays reviewed here do not provide specific information on certain genes and activated pathways, but rather reflect the global response toward radiation or drug treatment. Nonetheless, these techniques may offer the detection of predictive markers to identify individual radio- and chemosensitivity (90). Indeed, especially patients with genomic instability syndromes and deficiencies in specific proteins involved in DDR are predisposed to neoplastic diseases and show increased side effects during cancer treatment. Consequently, detection of hypersensitive patients is one promising approach of DDR assays (53,111).
DDR diagnostics have been successfully used to assess the efficacy of radiation and many DNA-damaging chemotherapeutic drugs. The commonly accepted concept is that quantitative DNA damage endpoints can be correlated with the clinical outcome of patients treated with genotoxic drugs (5). However, the introduction thereof into clinical routine has so far been hampered to a substantial extent by the lack of standardization and automation. Consequently, promising attempts have been reported recently to address these shortcomings and provide better conditions for the translation of these valuable tools into routine diagnostics (76,120). Additionally, new approaches based on plasmid reporters have been established for the assessment of individual DNA repair capacity covering multiple DNA damage repair pathways (121). Further, the development of new methods for liquid tumor biopsy like the isolation of circulating tumor cells or novel techniques to cultivate tumor cells, e.g., as 3D organoid cultures or spheroid systems have paved the way for the discovery of new DNA damage and DDR biomarkers (122,123).
Conclusions and perspective
Identification of a biomarker and its translation into routine clinical applications is a long and highly complex process, if it succeeds at all. The benefit of a marker needs to be proven by adequately powered, well-designed clinical trials considering various ethnological aspects as well as gender-, age- or tissue-specific variations. To meet the requirements of precision medicine as biomarker-based stratification of patients, potential novel assay technologies must be proven as robust and valid methods in the environment of modern laboratories. Thus, methods for precision DDR diagnostic such as the γH2AX or comet assays shall have to demonstrate that their level of automation and standardization is sufficient to be assessed in clinical studies. Such trials within the next five years can help to gain more clarity regarding the usefulness of DNA damage diagnostics and the readiness of their detection technologies for precision medicine.
Combined studies with other promising diagnostic tools like next generation sequencing need to demonstrate whether DNA damage assessment or DDR analysis provides additional, valuable information for diagnosis, prognosis or prediction of diseases in the context of precision medicine. If trials are successful, these findings may help to predict genome instability, cancer susceptibility or drug resistance and, thus, DNA damage analysis would support the development of tailored therapy regimes.
For DNA damage diagnostics, novel technologies have been developed to facilitate sample preparation and automated sample analysis. High-throughput analysis prospectively allows higher powered, standardized studies which hopefully soon provide distinct information on DNA damage or DDR markers in relation to their clinical application. Several research groups work on computer-assisted image interpretation platforms and first commercial systems are already available to conduct appropriate studies. These intriguing developments will provide new experience and gain valuable data to move forward in finding and establishing DDR biomarkers for personalized medicine. DNA damage and corresponding DDRs are fundamental events especially in cancer and identification of related biomarkers to predict treatment effectiveness and to reduce side effects will not only help patients but also safe time and costs of healthcare systems.
Acknowledgments
This work has in part been funded by the “Gesundheitscampus Brandenburg - Konsequenzen der alterassoziierten Zell- und Organfunktionen” initiative of the Brandenburgian Ministry of Science, Research and Culture (MWFK) and the InnoProfile-Transfer 03 IPT 611X project of the Federal Ministry of Education and Research (BMBF).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “DNA Damage Assessment for Precision Medicine”. The article has undergone external peer review.
Conflicts of Interest: The series “DNA Damage Assessment for Precision Medicine” was commissioned by the editorial office without any funding or sponsorship. D Roggenbuck is a shareholder of GA Generic Assays GmbH and MEDIPAN GmbH being diagnostic manufacturers. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lippi G, Plebani M. Personalized medicine: moving from simple theory to daily practice. Clin Chem Lab Med 2015;53:959-960. [Crossref] [PubMed]
- Schork NJ. Personalized medicine: Time for one-person trials. Nature 2015;520:609-611. [Crossref] [PubMed]
- Carlson B. What the devil is personalized medicine? Biotechnol Healthc 2008;5:17-19. [PubMed]
- Cirkel GA, Gadellaa-van Hooijdonk CG, Koudijs MJ, et al. Tumor heterogeneity and personalized cancer medicine: are we being outnumbered? Future Oncol 2014;10:417-428. [Crossref] [PubMed]
- Unger FT, Witte I, David KA. Prediction of individual response to anticancer therapy: historical and future perspectives. Cell Mol Life Sci 2015;72:729-57. [Crossref] [PubMed]
- US Food and Drug Administration. Guidance for Industry and Food and Drug Administration Staff. In Vitro Companion Diagnostic Devices. 2014. Available online: https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm077272.pdf
- Taube SE, Jacobson JW, Lively TG. Cancer diagnostics: Decision criteria for marker utilization in the clinic. Am J Pharmacogenomics 2005;5:357-364. [Crossref] [PubMed]
- Müller WU. Comet Assay. In: Obe G, Vijayalaxmi, editors. Chromosom Alterations - Methods, Results Importance Hum Heal With. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007.
- Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids 2011;2011:1-9.
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature 2001;411:366-74. [Crossref] [PubMed]
- Shaheen M, Allen C, Nickoloff JA, et al. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood 2011;117:6074-6082. [Crossref] [PubMed]
- Mladenov E, Magin S, Soni A, et al. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: Cell cycle and proliferation-dependent regulation. Semin Cancer Biol 2016;37-38:51-64. [Crossref] [PubMed]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009;361:1475-85. [Crossref] [PubMed]
- Schuler N, Rübe CE. Accumulation of DNA Damage-Induced Chromatin Alterations in Tissue-Specific Stem Cells: The Driving Force of Aging? PLoS One 2013;8:e63932 [Crossref] [PubMed]
- Phillips ER, McKinnon PJ. DNA double-strand break repair and development. Oncogene 2007;26:7799-808. [Crossref] [PubMed]
- Ambrose M, Gatti RA. Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions. Blood 2013;121:4036-4045. [Crossref] [PubMed]
- Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci 2014;105:370-88. [Crossref] [PubMed]
- Miyakoshi J. Cellular and Molecular Responses to Radio-Frequency Electromagnetic Fields. Proc IEEE 2013;101:1494-1502. [Crossref]
- Kumari S, Rastogi RP, Singh KL, et al. DNA Damage: Detection Strategies. EXCLI J 2008;7:44-62.
- Gunasekarana V, Raj GV, Chand P. A comprehensive review on clinical applications of comet assay. J Clin Diagn Res 2015;9:GE01-5. [PubMed]
- Agrawala PK, Adhikari JS, Chaudhury NK. Lymphocyte chromosomal aberration assay in radiation biodosimetry. J Pharm Bioallied Sci 2010;2:197-201. [Crossref] [PubMed]
- Rothkamm K, Beinke C, Romm H, et al. Comparison of established and emerging biodosimetry assays. Radiat Res 2013;180:111-9. [Crossref] [PubMed]
- Schmid TE, Zlobinskaya O, Multhoff G. Differences in Phosphorylated Histone H2AX Foci Formation and Removal of Cells Exposed to Low and High Linear Energy Transfer Radiation. Curr Genomics 2012;13:418-25. [Crossref] [PubMed]
- Suspiro A, Prista J. Biomarkers of occupational exposure do anticancer agents: A minireview. Toxicol Lett 2011;207:42-52. [Crossref] [PubMed]
- M’kacher R, Maalouf EEL, Ricoul M, et al. New tool for biological dosimetry: reevaluation and automation of the gold standard method following telomere and centromere staining. Mutat Res 2014;770:45-53. [Crossref] [PubMed]
- Dearfield KL, Thybaud V, Cimino MC, et al. Follow-up actions from positive results of in vitro genetic toxicity testing. Environ Mol Mutagen 2011;52:177-204. [Crossref] [PubMed]
- Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc 2007;2:1084-104. [Crossref] [PubMed]
- Mateuca R, Lombaert N, Aka PV, et al. Chromosomal changes: induction, detection methods and applicability in human biomonitoring. Biochimie 2006;88:1515-31. [Crossref] [PubMed]
- Rosefort C, Fauth E, Zankl H. Micronuclei induced by aneugens and clastogens in mononucleate and binucleate cells using the cytokinesis block assay. Mutagenesis 2004;19:277-84. [Crossref] [PubMed]
- Hashimoto K, Nakajima Y, Matsumura S, et al. An in vitro micronucleus assay with size-classified micronucleus counting to discriminate aneugens from clastogens. Toxicol In Vitro 2010;24:208-216. [Crossref] [PubMed]
- Musgrove C, Camps M. Models for Detection of Genotoxicity in vivo: Present and Future. Biochem Genet Mol Biol 2012. p. 31-50.
- Pinto MM, Santos NF, Amaral A. Current status of biodosimetry based on standard cytogenetic methods. Radiat Environ Biophys 2010;49:567-81. [Crossref] [PubMed]
- Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006;1:23-29. [Crossref] [PubMed]
- Collins AR, Oscoz AA, Brunborg G, et al. The comet assay: Topical issues. Mutagenesis 2008;23:143-51. [Crossref] [PubMed]
- Azqueta A, Shaposhnikov S, Collins AR. DNA oxidation: Investigating its key role in environmental mutagenesis with the comet assay. Mutat Res 2009;674:101-8. [Crossref] [PubMed]
- Shaposhnikov S, Thomsen PD, Collins AR. Combining fluorescent in situ hybridization with the comet assay for targeted examination of DNA damage and repair. Methods Mol Biol 2011;682:115-32. [Crossref] [PubMed]
- Garcia-Canton C, Anadón A, Meredith C. γH2AX as a novel endpoint to detect DNA damage: applications for the assessment of the in vitro genotoxicity of cigarette smoke. Toxicol In Vitro 2012;26:1075-86. [Crossref] [PubMed]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858-5868. [Crossref] [PubMed]
- Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer 2008;8:957-967. [Crossref] [PubMed]
- Koch U, Höhne K, Von Neubeck C, et al. Residual γH2AX foci predict local tumour control after radiotherapy. Radiother Oncol 2013;108:434-9. [Crossref] [PubMed]
- Menegakis A, De Colle C, Yaromina A, et al. Residual γH2AX foci after ex vivo irradiation of patient samples with known tumour-type specific differences in radio-responsiveness. Radiother Oncol 2015;116:480-5. [Crossref] [PubMed]
- Löbrich M, Shibata A, Beucher A, et al. γH2AX foci analysis for monitoring DNA double-strand break repair: Strengths, limitations and optimization. Cell Cycle 2010;9:662-9. [Crossref] [PubMed]
- Nakamura AJ, Rao VA, Pommier Y, et al. The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle 2010;9:389-397. [Crossref] [PubMed]
- Polo SE. Reshaping Chromatin after DNA Damage: The Choreography of Histone Proteins. J Mol Biol 2015;427:626-36. [Crossref] [PubMed]
- Sak A, Stuschke M. Use of γH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin Radiat Oncol 2010;20:223-31. [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Sordet O, et al. γ-H2AX Detection in Peripheral Blood Lymphocytes, Splenocytes, Bone Marrow, Xenografts, and Skin. Methods Mol Biol 2011;682:249-70. [Crossref] [PubMed]
- Sak A, Grehl S, Erichsen P, et al. γ-H2AX foci formation in peripheral blood lymphocytes of tumor patients after local radiotherapy to different sites of the body: Dependence on the dose-distribution, irradiated site and time from start of treatment. Int J Radiat Biol 2007;83:639-52. [Crossref] [PubMed]
- Andrievski A, Wilkins RC. The response of γ-H2AX in human lymphocytes and lymphocytes subsets measured in whole blood cultures. Int J Radiat Biol 2009;85:369-376. [Crossref] [PubMed]
- Lisowska H, Wegierek-Ciuk A, Banasik-Nowak A, et al. The dose-response relationship for dicentric chromosomes and γ-H2AX foci in human peripheral blood lymphocytes: Influence of temperature during exposure and intra- and inter-individual variability of donors. Int J Radiat Biol 2013;89:191-9. [Crossref] [PubMed]
- Martin OA, Ivashkevich A, Choo S, et al. Statistical analysis of kinetics, distribution and co-localisation of DNA repair foci in irradiated cells: Cell cycle effect and implications for prediction of radiosensitivity. DNA Repair (Amst) 2013;12:844-55. [Crossref] [PubMed]
- Horn S, Barnard S, Brady D, et al. Combined analysis of gamma-H2AX/53BP1 foci and caspase activation in lymphocyte subsets detects recent and more remote radiation exposures. Radiat Res 2013;180:603-9. [Crossref] [PubMed]
- Rothkamm K, Horn S. gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita 2009;45:265-271. [PubMed]
- Rübe CE, Fricke A, Schneider R, et al. DNA repair alterations in children with pediatric malignancies: novel opportunities to identify patients at risk for high-grade toxicities. Int J Radiat Oncol Biol Phys 2010;78:359-69. [Crossref] [PubMed]
- Lorat Y, Schanz S, Schuler N, et al. Beyond repair foci: DNA double-strand break repair in euchromatic and heterochromatic compartments analyzed by transmission electron microscopy. PLoS One 2012;7:e38165 [Crossref] [PubMed]
- Lorat Y, Brunner CU, Schanz S, et al. Nanoscale analysis of clustered DNA damage after high-LET irradiation by quantitative electron microscopy - The heavy burden to repair. DNA Repair (Amst) 2015;28:93-106. [Crossref] [PubMed]
- Rödiger S, Schneider J, Sauer L, et al. A Multiparameter Platform for the Analysis of DNA Double-strand Breaks and Associated Biomarkers after Genome Editing. F1000Research 2016;5:350. (poster).
- Sauer L, Rödiger S, Schneider J, et al. Functional Multiparameter Analysis of Double-Strand Breaks and Associate Biomarkers during Genome Editing. F1000Research 2016;5:706. (poster).
- Nikolova T, Dvorak M, Jung F, et al. The γH2AX assay for genotoxic and nongenotoxic agents: comparison of H2AX phosphorylation with cell death response. Toxicol Sci 2014;140:103-17. [Crossref] [PubMed]
- Schunck C, Johannes T, Varga D, et al. New developments in automated cytogenetic imaging: Unattended scoring of dicentric chromosomes, micronuclei, single cell gel electrophoresis, and fluorescence signals. Cytogenet Genome Res 2004;104:383-9. [Crossref] [PubMed]
- Romm H, Ainsbury E, Barnard S, et al. Automatic scoring of dicentric chromosomes as a tool in large scale radiation accidents. Mutat Res 2013;756:174-83. [Crossref] [PubMed]
- M’kacher R, El Maalouf E, Terzoudi G, et al. Detection and automated scoring of dicentric chromosomes in nonstimulated lymphocyte prematurely condensed chromosomes after telomere and centromere staining. Int J Radiat Oncol Biol Phys 2015;91:640-9. [Crossref] [PubMed]
- Varga D, Johannes T, Jainta S, et al. An automated scoring procedure for the micronucleus test by image analysis. Mutagenesis 2004;19:391-7. [Crossref] [PubMed]
- Frieauff W, Martus HJ, Suter W, et al. Automatic analysis of the micronucleus test in primary human lymphocytes using image analysis. Mutagenesis 2013;28:15-23. [Crossref] [PubMed]
- Rodrigues MA, Beaton-Green LA, Kutzner BC, et al. Automated analysis of the cytokinesis-block micronucleus assay for radiation biodosimetry using imaging flow cytometry. Radiat Environ Biophys 2014;53:273-82. [Crossref] [PubMed]
- Fenech M, Kirsch-Volders M, Rossnerova A, et al. HUMN project initiative and review of validation, quality control and prospects for further development of automated micronucleus assays using image cytometry systems. Int J Hyg Environ Health 2013;216:541-52. [Crossref] [PubMed]
- Ge J, Prasongtanakij S, Wood DK, et al. CometChip: A High-throughput 96-Well Platform for Measuring DNA Damage in Microarrayed Human Cells. J Vis Exp 2014;e50607 [PubMed]
- Watson C, Ge J, Cohen J, et al. High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using cometchip technology. ACS Nano 2014;8:2118-33. [Crossref] [PubMed]
- Böcker W, Rolf W, Bauch T, et al. Automated comet assay analysis. Cytometry 1999;35:134-44. [Crossref] [PubMed]
- Ritter D, Knebel J. Genotoxicity testing in vitro - development of a higher throughput analysis method based on the comet assay. Toxicol In Vitro 2009;23:1570-5. [Crossref] [PubMed]
- Collins A, Koppen G, Valdiglesias V, et al. The comet assay as a tool for human biomonitoring studies: The ComNet Project. Mutat Res Rev Mutat Res 2014;759:27-39. [Crossref] [PubMed]
- Koppen G, Azqueta A, Pourrut B, et al. The next three decades of the comet assay: a report of the 11th International Comet Assay Workshop. Mutagenesis 2017;32:397-408. [Crossref] [PubMed]
- Valente M, Voisin P, Laloi P, et al. Automated gamma-H2AX focus scoring method for human lymphocytes after ionizing radiation exposure. Radiat Meas 2011;46:871-6. [Crossref]
- Ivashkevich AN, Martin OA, Smith AJ, et al. γH2AX foci as a measure of DNA damage: A computational approach to automatic analysis. Mutat Res 2011;711:49-60. [Crossref] [PubMed]
- Oeck S, Malewicz NM, Hurst S, et al. The Focinator - a new open-source tool for high-throughput foci evaluation of DNA damage. Radiat Oncol 2015;10:163. [Crossref] [PubMed]
- Lapytsko A, Kollarovic G, Ivanova L, et al. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics 2015;16:392. [Crossref] [PubMed]
- Runge R, Hiemann R, Wendisch M, et al. Fully automated interpretation of ionizing radiation-induced γH2AX foci by the novel pattern recognition system AKLIDES®. Int J Radiat Biol 2012;88:439-447. [Crossref] [PubMed]
- Hernández L, Terradas M, Martín M, et al. Highly sensitive automated method for DNA damage assessment: gamma-H2AX foci counting and cell cycle sorting. Int J Mol Sci 2013;14:15810-15826. [Crossref] [PubMed]
- Turner HC, Brenner DJ, Chen Y, et al. Adapting the γ-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res 2011;175:282-90. [Crossref] [PubMed]
- Hiemann R, Büttner T, Krieger T, et al. Challenges of automated screening and differentiation of non-organ specific autoantibodies on HEp-2 cells. Autoimmun Rev 2009;9:17-22. [Crossref] [PubMed]
- Rödiger S, Schierack P, Böhm A, et al. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. Adv Biochem Eng Biotechnol 2013;133:35-74. [Crossref] [PubMed]
- Sowa M, Hiemann R, Schierack P, et al. Next-Generation Autoantibody Testing by Combination of Screening and Confirmation—the CytoBead® Technology. Clin Rev Allergy Immunol 2017;53:87-104. [Crossref] [PubMed]
- Willitzki A, Lorenz S, Hiemann R, et al. Fully automated analysis of chemically induced gammaH2AX foci in human peripheral blood mononuclear cells by indirect immunofluorescence. Cytometry A 2013;83:1017-1026. [Crossref] [PubMed]
- Leifert WR, Siddiqui SM. γH2AX is a biomarker of modulated cytostatic drug resistance. Cytometry A 2015;87:692-695. [Crossref] [PubMed]
- Apostolou P, Toloudi M, Kourtidou E, et al. Use of the comet assay technique for quick and reliable prediction of in vitro response to chemotherapeutics in breast and colon cancer. J Biol Res (Thessalon) 2014;21:14. [Crossref] [PubMed]
- Guillerman RP. From ‘Image Gently’ to image intelligently: a personalized perspective on diagnostic radiation risk. Pediatr Radiol 2014;44:444-9. [Crossref] [PubMed]
- Skripcak T, Belka C, Bosch W, et al. Creating a data exchange strategy for radiotherapy research: towards federated databases and anonymised public datasets. Radiother Oncol 2014;113:303-9. [Crossref] [PubMed]
- Ziegler A, Koch A, Krockenberger K, et al. Personalized medicine using DNA biomarkers: A review. Hum Genet 2012;131:1627-38. [Crossref] [PubMed]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071-8. [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Martin OA, et al. Recent developments in the use of γ-H2AX as a quantitative DNA double-strand break biomarker. Aging (Albany NY) 2011;3:168-174. [Crossref] [PubMed]
- Redon CE, Weyemi U, Parekh PR, et al. γ-H2AX and other histone post-translational modifications in the clinic. Biochim Biophys Acta 2012;1819:743-56.
- Kulka U, Ainsbury L, Atkinson M, et al. Realising the European network of biodosimetry: RENEB-status quo. Radiat Prot Dosimetry 2015;164:42-45. [Crossref] [PubMed]
- Jaworska A, Ainsbury EA, Fattibene P, et al. Operational guidance for radiation emergency response organisations in Europe for using biodosimetric tools developed in eu multibiodose project. Radiat Prot Dosimetry 2015;164:165-169. [Crossref] [PubMed]
- Barnard S, Ainsbury EA, Al-Hafidh J, et al. The first gamma-H2AX biodosimetry intercomparison exercise of the developing european biodosimetry network RENEB. Radiat Prot Dosimetry 2015;164:265-270. [Crossref] [PubMed]
- Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A 2003;100:5057-62. [Crossref] [PubMed]
- Kuefner MA, Brand M, Engert C, et al. Radiation Induced DNA Double-Strand Breaks in Radiology. RoFo 2015;187:872-878. [Crossref] [PubMed]
- Habash M, Bohorquez LC, Kyriakou E, et al. Clinical and functional assays of radiosensitivity and radiation-induced second cancer. Cancers (Basel) 2017;9:1-18. [Crossref] [PubMed]
- McKenna DJ, McKeown SR, McKelvey-Martin VJ. Potential use of the comet assay in the clinical management of cancer. Mutagenesis 2008;23:183-90. [Crossref] [PubMed]
- Siddiqui MS, François M, Fenech MF, et al. Persistent γH2AX: A promising molecular marker of DNA damage and aging. Mutat Res Rev Mutat Res 2015;766:1-19. [Crossref] [PubMed]
- Djuzenova CS, Schindler D, Stopper H, et al. Identification of ataxia telangiectasia heterozygotes, a cancer-prone population, using the single-cell gel electrophoresis (Comet) assay. Lab Invest 1999;79:699-705. [PubMed]
- Bürger S, Schindler D, Fehn M, et al. Radiation-induced DNA damage and repair in peripheral blood mononuclear cells from Nijmegen breakage syndrome patients and carriers assessed by the Comet assay. Environ Mol Mutagen 2006;47:260-70. [Crossref] [PubMed]
- Schuler N, Palm J, Kaiser M, et al. DNA-damage foci to detect and characterize DNA repair alterations in children treated for pediatric malignancies. PLoS One 2014;9:e91319 [Crossref] [PubMed]
- Pouliliou S, Koukourakis MI. Gamma histone 2AX (γ-H2AX)as a predictive tool in radiation oncology. Biomarkers 2014;19:167-80. [Crossref] [PubMed]
- Willers H, Gheorghiu L, Liu Q, et al. DNA Damage Response Assessments in Human Tumor Samples Provide Functional Biomarkers of Radiosensitivity. Semin Radiat Oncol 2015;25:237-50. [Crossref] [PubMed]
- McKeown SR, Robson T, Price ME, et al. Potential use of the alkaline comet assay as a predictor of bladder tumour response to radiation. Br J Cancer 2003;89:2264-70. [Crossref] [PubMed]
- Lacombe J, Azria D, Mange A, et al. Proteomic approaches to identify biomarkers predictive of radiotherapy outcomes. Expert Rev Proteomics 2013;10:33-42. [Crossref] [PubMed]
- Lage H. An overview of cancer multidrug resistance: A still unsolved problem. Cell Mol Life Sci 2008;65:3145-67. [Crossref] [PubMed]
- Mattii L, Barale R, Petrini M. Use of the comet test in the evaluation of multidrug resistance of human cell lines. Leukemia 1998;12:627-32. [Crossref] [PubMed]
- Zhou F, Mei H, Wu Q, et al. Expression of histone H2AX phosphorylation and its potential to modulate adriamycin resistance in K562/A02 cell line. J Huazhong Univ Sci Technolog Med Sci 2011;31:154-8. [Crossref] [PubMed]
- Reddig A, Lorenz S, Hiemann R, et al. Assessment of modulated cytostatic drug resistance by automated γH2AX analysis. Cytometry A 2015;87:724-32. [Crossref] [PubMed]
- Mah LJ, El-Osta A, Karagiannis TC. GammaH2AX as a molecular marker of aging and disease. Epigenetics 2010;5:129-36. [Crossref] [PubMed]
- Valdiglesias V, Giunta S, Fenech M, et al. γH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat Res 2013;753:24-40. [Crossref] [PubMed]
- Milic M, Frustaci A, Del Bufalo A, et al. DNA damage in non-communicable diseases: A clinical and epidemiological perspective. Mutat Res 2015;776:118-27. [Crossref] [PubMed]
- Lippi G, Buonocore R, Tarperi C, et al. DNA injury is acutely enhanced in response to increasing bulks of aerobic physical exercise. Clin Chim Acta 2016;460:146-151. [Crossref] [PubMed]
- Davison GW. Exercise and oxidative damage in nucleoid DNA quantified using single cell gel electrophoresis: Present and future application. Front Physiol 2016;7:249. [PubMed]
- Danese E, Lippi G, Sanchis-Gomar F, et al. Physical Exercise and DNA Injury: Good or Evil? Adv Clin Chem 2017;81:193-230. [Crossref] [PubMed]
- Reddig A, Fatahi M, Friebe B, et al. Analysis of DNA Double-Strand Breaks and Cytotoxicity after 7 Tesla Magnetic Resonance Imaging of Isolated Human Lymphocytes. PLoS One 2015;10:e0132702 [Crossref] [PubMed]
- Fatahi M, Reddig A. DNA double-strand breaks and micronuclei in human blood lymphocytes after repeated whole body exposures to 7T Magnetic Resonance Imaging. Neuroimage 2016;133:288-93. [Crossref] [PubMed]
- Reddig A, Fatahi M, Roggenbuck D, et al. Impact of in Vivo High-Field-Strength and Ultra-High-Field-Strength MR Imaging on DNA Double-Strand-Break Formation in Human Lymphocytes. Radiology 2017;282:782-9. [Crossref] [PubMed]
- Danese E, Lippi G, Buonocore R, et al. Mobile phone radiofrequency exposure has no effect on DNA double strand breaks (DSB) in human lymphocytes. Ann Transl Med 2017;5:272. [Crossref] [PubMed]
- Liu SK, Olive PL, Bristow RG. Biomarkers for DNA DSB inhibitors and radiotherapy clinical trials. Cancer Metastasis Rev 2008;27:445-58. [Crossref] [PubMed]
- Nagel ZD, Margulies CM, Chaim IA, et al. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc Natl Acad Sci 2014;111:E1823-32. [Crossref] [PubMed]
- Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng Part B Rev 2008;14:61-86. [Crossref] [PubMed]
- Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng 2009;103:655-63. [Crossref] [PubMed]
Cite this article as: Reddig A, Rübe CE, Rödiger S, Schierack P, Reinhold D, Roggenbuck D. DNA damage assessment and potential applications in laboratory diagnostics and precision medicine. J Lab Precis Med 2018;3:31.


