Aberrant non-coding RNA expression profiles as biomarker/bio-signature in autoimmune and inflammatory rheumatic diseases
Introduction
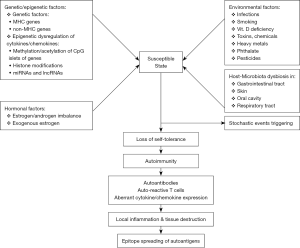
Autoimmune status is elicited by multi-etiologic factors (1,2) that may include genetic predisposition (3-5), epigenetic dysregulation (5-7), gender bias and hormone imbalance (8-11), environmental stimulation (12,13), host-microbiota dysbiosis (14-18) and triggering from stochastic events (1,2,19). The intricate interactions among these etiologic factors lead to a state of “loss of self-tolerance” in susceptible individuals. Furthermore, a self-sustaining mechanism operates via autoimmune-mediated local inflammation, tissue destruction, and autoantigen epitope spreading elicited by sophisticated interactions among pathogenic autoantibodies, autoreactive T cells and pro-inflammatory cytokines (1-21). A scheme demonstrating the involvement of these factors in the pathogenesis of autoimmunity is shown in Figure 1.
In the respect of epigenetic modulation of immunity and autoimmunity (22-24), there are many genetic on/off regulatory modes such as methylation/acetylation of CpG islets in cytokine genes (7,25-27), histone modification by histone deacetylase/histone acetyltransferase (22,23,28-30) and post-transcriptional modification of messenger RNAs (mRNAs) by non-coding RNA (ncRNA) (31-36). On the other hand, high-throughput detection technology of transcriptomes and bioinformatic analysis have found the presence of thousands ncRNAs in the cytoplasm and body fluids in charge of regulating mRNA expression. In conjunction with proteomic (37-39) and metabolomic profiling technologies (40-42), many investigations have tried to find the useful biomarkers/bio-signatures for diagnosis, disease activity & therapeutic monitoring as well as outcome prediction of autoimmune and inflammatory rheumatic diseases. In fact, the “omics” studies have demonstrated many biomarkers or bio-signatures in the literature (43-46). In addition, these array profiles can concomitantly elucidate the potential molecular pathways in the development of autoimmune and inflammatory rheumatic diseases (44). However, either biomarkers or bio-signatures must fulfill the criteria (47,48) as shown in Table 1 to become useful monitoring parameters in the clinical practice.
Table 1
| Definition of biomarker by the Biomarker Definition Working Group |
| “A characteristic that is objectively measured and evaluated as an indicator or normal biological processes, pathological processes, or pharmacological responses to a therapeutic intervention” |
| Definition of bio-signature |
| The obtained data from high-throughput “omics” together define a biomarker |
| A major issue is the low reproducibility and limited biological interpretability of the candidate biomarker signature |
| The detected molecular species include genes and their transcripts, proteins, metabolites and non-coding regulatory RNAs |
| Types of biomarkers |
| Predictive (risk) biomarkers |
| Diagnostic biomarkers |
| Disease activity monitoring biomarker |
| Prognostic biomarkers |
| Ideal criteria for a biomarker |
| A disease-causing molecule with high sensitivity and specificity |
| General usability and high reproducibility |
| Low cost |
| Logistic interpretability |
The ncRNAs are conventionally classified into two categories, long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs), cutoff at the nucleotide (nt) or base pair (bp) number of 200. The RNAs with molecular size larger than 200 nt are classified as lncRNAs whereas those with molecular size shorter than 200 nt belong to the sncRNA category. lncRNAs can be further divided into 7 subtypes according to their lineage-specific effects on mRNA regulation for the innate and adaptive immune homeostasis (34,35) (Figure 2). In contrast, sncRNAs are divided into at least nine subtypes according to the size, argonaute (Ago) protein association and their major localization (Table 2) (36,49). Single nucleotide polymorphisms (SNP) in human genes disturb genome stability (49) and induce inflammatory rheumatic disorders (35). In this review, we will discuss in detail the biological functions of ncRNAs in the development of immune system and their aberrant expression profiles resulting in the pathogenetic and pathological processes of various autoimmune-related diseases.
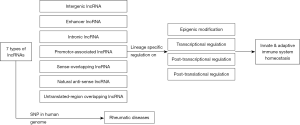
Table 2
| Nomenclature of ncRNA | Size (bp) | Ago protein association | Localization |
|---|---|---|---|
| Long non-coding RNAs (lncRNA) | >200 | N/A | Nucleus |
| Small non-coding RNAs (sncRNA) | <200 | Ago | Nucleus, cytoplasm |
| Small interference RNAs (siRNA) & endogenous siRNA | 20–24 | Ago | Nucleus |
| Guide RNAs (gRNA) | 50–70 | N/A | Nucleus |
| PIWI interacting RNAs (piRNA) | 23–31 | PIWI | Nucleus, cytoplasm |
| Promotor association RNAs (pRNA) | Nucleus | ||
| Small nucleolar RNAs (snoRNA) & sno-derived RNAs | <200 | Nucleolus | |
| MicroRNAs (miRNA) | 20–24 | Ago | Cytoplasm |
| Double-stranded break-induced small RNAs (diRNA) | 20–24 | Ago | Nucleus |
| Circular RNAs (cirRNA) | Nucleus | ||
| Exosomal miRNAs (exo-miR) | 20–24 | Ago | Plasma, body fluids, extracellular space |
Ago, argonaute; PIWI, P-element-induced wimpy testis.
Biology of ncRNAs
RNAs are traditionally regarded as informational intermediate between a DNA (gene) and its encoding product protein. In fact, around 70–85% of the human genome are actively transcribed into RNAs. However, only 2% are transcribed into protein-coding mRNAs. This fact implies that the number of ncRNAs is much higher than that of the protein coding genes (36,50). lncRNAs (>200 bp) are lineage-specific regulators of mRNAs, which can modulate innate and adaptive immune homeostasis by epigenetic, transcriptional, post-transcriptional and post-translational regulations (Figure 2). In contrast, miRNA (20–24 bp in length) can target several transcripts rather than a single specific transcript of genes in the site of 3'-UTR (51). Up to the present, more than 9,000 miRNAs have been identified to carry out various enhancing or suppressing functions on mRNA (52,53). These modulatory functions of miRNAs are obviously crucial in physiological and pathological conditions (36). Figure 3 demonstrates the modulation of miRNAs on the cell cycle, cell differentiation and cell apoptosis by way of different inhibitory activities on mRNA. Aberrant expression of ncRNAs may induce a number of autoimmune, inflammatory rheumatic, and neoplastic diseases. Indeed, an autoimmune disease is mainly characterized by the presence of autoantibodies and autoreactive T lymphocytes. Examples include systemic lupus erythematosus (SLE) or type 1 diabetes mellitus (T1DM). On the other hand, an inflammatory rheumatic disease is characterized mainly by inflammation rather than a presence of obvious autoantibodies or autoreactive cells. These include seronegative spondylarthropathy or inflammatory bowel disease (IBDs).

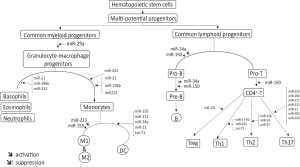
miRNAs are crucial regulators in the development of hematopoietic stem cells and immune systems
It has been demonstrated that the differentiation and homeostasis of the hematopoietic system require complex interconnected regulatory networks to distinguish the different blood cell lineages. The major immune system includes innate (monocytes, macrophages, dendritic cells, natural killers and leukocytes) and adaptive (B cells, T cells and a variety of T cell subsets) immune cells, which are originated from hematopoietic stem cells. Among the regulatory molecules in hematopoiesis, miRNAs play a pivotal role in the fine-tuning of differentiation in the system (31,33,54,55). Knockout or silencing of certain miRNA machinery results in severe compromise of the immune system. The involvement of miRNAs in immune system development is depicted in Figure 4. The aberrant expression of miRNAs in hematopoiesis can undoubtedly elicit autoimmune, inflammatory rheumatic and neoplastic diseases (56-58).
Aberrant miRNA expression profiles of T cells become bio-signature of the pathogenesis and disease activity in patients with SLE
Many investigators have tried to detect and confirm the miRNA expression profiles in the immune cells, plasma or other body fluids by using miRNA extraction kits, miRNA reverse transcription kits and miRNA microarray for early detection, and real-time quantitative polymerase chain reaction (PCR) for confirmation. However, the data of individual T cell miRNA expression profiles in the literature for SLE risk or pathogenesis are quite variable (44,59-67). Lu et al. (62,65) have found decreased miR-145, increased miR-224, and aberrant Ca2+ influx-regulated ncRNAs play roles in lupus pathogenesis. Later, they have extensively reviewed the literature and concluded that a number of elevated miRNAs could potentially become bio-signatures for immunopathogenesis of SLE (68). These bio-signatures include elevation of miR-17–92 cluster, miR-21, miR-296, miR-126, miR-148a, miR-224, miR-524-5p, and suppression of miR-31, miR-125a, miR-125b, miR-142-3p, miR-142-5p and miR-146a. In addition, these bio-signatures are found intriguingly correlated with T cell subset alteration, aberrant cytokine/chemokine release, altered gene transcription and immune cell signaling abnormalities in SLE (68). Besides, urinary exosomal miRNA profiling was also investigated as bio-signatures for lupus nephritis (69-71). These include increased miR-125a, miR-146, miR-150 and miR-155, and decreased miR-141, miR-192 and miR-200a. For exploring the miRNA expression profiles in the damaged target tissues, Cardenas-Gonzalez et al. (72) directly identified, confirmed and explicated miR-30c-5p, miR-1273e and miR-3201 in the renal tissue of patients with lupus nephritis. This cause-effect relationship investigation of the damaged tissue is direct and more reliable than the conventional correlation analysis (73,74).
Abnormal miRNA expression profile reflects the pathogenesis, helps diagnosis and indicates therapeutic prognosis in patients with rheumatoid arthritis (RA)
Similar to the miRNA study in SLE, different investigators have demonstrated variable miRNA expression profiles in patients with RA, reflecting its pathogenesis and helping therapeutic monitoring (74-82). The abnormal miRNA expression in RA includes increase in miR-15a, miR-16, miR-21, miR-25, miR-124a, miR-146a, miR-155, miR-203, miR-223 and miR-346, and decrease in miR-140-3p and miR-140-5p. Some unique therapeutic strategy, using miRNA antagonist or agonist, can ameliorate the inflammation in RA (83-85). Shi et al (83) found miR-27a could inhibit migration and invasion of fibroblast-like synoviocytes by targeting follistatin-like protein 1 in RA (83). Sharma et al. (79) demonstrated key components of cytokine signaling and inflammation which is regulated by miRNA. Furthermore, Lai et al. (85) have found that anti-citrullinated protein antibodies can suppress let-7a expression and facilitate inflammatory responses in patients with RA. Lai et al. (68) have tried to correlate the miRNA expression profile with rheumatoid pathogenesis by meta-analysis. They noted a decrease in miR-21 expression, enhanced STAT3 but suppressed STAT5, which upsurge T-helper 17 (Th17)/regulatory T cell (Treg) ratio. Increased expression of miR-23 may diminish IL-10 production, leading to imbalance between pro-inflammatory and anti-inflammatory cytokine production. Furthermore, increased LOC100506036 (a kind of lncRNAs) expression enhanced transcription factors such as nuclear factor of activated T cell (NFAT) and Smith deoxyribonuclease protein (SMDP), which eventually activates T cells.
Aberrant miRNA transcription in other systemic and organ-specific autoimmune diseases
Primary Sjögren’s syndrome (pSS)
pSS is featured by systemic autoimmunity and chronic inflammation with dysfunction of exocrine glands. Twenty-five miRNAs including miR-146a, miR-16 and miR-21 were found over-expressed in both pSS and SLE patients. On the contrary, down-regulation of miR-150-5p, which is novel and unique, has been found in pSS (86,87). Wang-Renault et al. (88) further demonstrated that hsa-miR-30b-5p, hsa-miR-222-3p, hsa-miR-26a-5p, hsa-miR-30b-5p and hsa-miR-19b-3p were differentially expressed in B cells of pSS patients. Functional studies revealed that inhibition of hsa-miR-30b-5p by miRNA antagonist enhanced the expression of B cell activating factor of TNFR superfamily (BAFF) in B cells originated from pSS patients. These miRNA expression profiles can become the pathogenetic bio-signatures of pSS.
Anti-phospholipid syndrome (APS)
APS is diagnosed in autoimmune patients with a persistent presence of anti-phospholipid antibodies against mainly β2-glycoprotein I and different phospholipids including phosphatidylserine, phosphatidylcholine, and phosphatidylethanolamine, which manifests as arterial or venous thrombosis as well as pregnancy morbidity. The miRNA expression profiles relevant to APS were miR-19b and miR-20a that were implicated in the signaling pathways of TGF-β and vascular endothelial cell growth factor (VEGF), hypoxia and angiogenesis (89,90). These miRNAs can potentially be used as pathogenetic bio-signatures for primary APS. However, no investigation has been reported for the specific miRNA expression profile relevant to obstetric APS patients.
Systemic sclerosis (SSc)
SSc is characterized by Raynaud’s phenomenon in the early stage which eventually leads to generalized fibrosis of the skin and internal organs due to overproduction of TGF-β. Steen et al. (91) evaluated the cell-free miRNA expression profile in plasma from SSc patients. They found that miR-16, miR-223, and miR-638 were elevated and relevant to the TGF-β signaling and tissue fibrosis. In addition, miR-638 was found weakly correlated with the serum titer of anti-Scl-70 antibody. The authors also noted the differential expression of miR-142-3p, miR-150, miR-150, and miR-638 among SSc and SLE patients.
T1DM
T1DM is an organ-specific autoimmune disease characterized by selective destruction of pancreatic β-cells driven by the immune dysfunction. The role of miRNAs in T1DM has been explored by experiments using known pancreatic islets as cell-based disease models (92). Indeed, it was found that virus-induced miRNA dysregulation seemed implicated in the immune-mediated β-cell destruction. Zheng et al. (93) have reviewed recent data by focusing on the miRNAs involved in immune homeostasis and regulation of the β-cell function in T1DM. Assmann et al. (94) have also extensively reviewed the literature and found the results were inconclusive with only few miRNAs consistently dysregulated among these studies. It is concluded that 11 miRNAs including miR-21-5p, miR-24-3p, miR-100-5p, miR-146-5p, miR-148a-3p, miR-150-5p, miR-181a-5p, miR-210-5p, miR-342-3p, miR-375 and miR-1275 may potentially become the circulating pathogenetic bio-signatures for T1DM.
Myasthenia gravis (MG)
MG is characterized by the progressive muscle weakness and the presence of serum autoantibodies specific for either acetylcholine receptor (AChR+) or muscle-specific tyrosine kinase (MuSK+). These two autoantibodies are antagonistic antibodies that can block the neuronal transmission or muscle contraction. However, they do not reflect exactly the disease progression. Punga et al. (95) demonstrated that plasma levels of miR-150-5p and miR-21-5p were elevated in AchR+ MG patients who were immunosuppressed and improved clinically after thymoma resection. On the other hand, up-regulation of let-7 family has been found in MuSK+ MG patients. These circulating miRNAs can be considered as useful biomarkers for diagnoses, disease activity evaluation, therapeutic monitoring and foreseeing prognosis in MG patients.
Graves’ disease (GD)
GD is an archetype of organ-specific autoimmune disease characterized by aberrant Treg function and subsequent production of anti-thyroid stimulatory hormone receptor (TSHR) antibodies (96). The anti-TSHR antibodies are agonistic antibodies that can facilitate the synthesis and secretion of thyroxine. Hiratsuka et al. (97) demonstrated that increased let-78-3p and miR-339-5p as well as decreased miR-23b-5p and miR-92a-39 in intractable GD can lead to IL-1β and TNF-α production and suppression of the Treg function.
Multiple sclerosis (MS)
MS is a life-long organ-specific autoimmune inflammatory disorder of central nervous system featured by immune cell infiltration, degeneration of axons and neurons, local demyelination/remyelination and astrogliosis. Kacperska et al. (98) have reviewed the literature and concluded that circulating miR-146 and miR-153 are correlated with high sustenance, tissue specificity and TLR-4 activation (indicating inflammation) in MS patients.
Table 3 summarizes various ncRNA-mediated pathological processes in different autoimmune diseases.
Table 3
| Diseases | Bio-signature | Pathological process |
|---|---|---|
| Systemic lupus erythematosus | miR-21↑, miR-29b↑, miR-126↑ & miR-148a↑ | DNA hypomethylation (68) |
| miR-142-3p↓ & miR-142-5p↓ | T & B cell activation (68) | |
| miR-146a↓ | Type I IFN↑ (68) | |
| miR-224↑ | Cell apoptosis↑ (68) | |
| miR-21↑, miR-31↓, miR-142-3p↓ & miR-410↓ | IL-10↑ (68) | |
| miR-125a↓ & miR-125b↓ | Th17/Treg ratio↑ (68) | |
| miR1273e↓& miR-3201↓ | Endocapillary glomerular inflammation (72) | |
| Rheumatoid arthritis | LOC100506036↑ | T cell activation (68) |
| miR-223↑ | Pro/anti-inflammatory ratio↑ & cytokine imbalance (68) | |
| miR-21↓ | Th17/Treg ratio↑ (68) | |
| Sjögren’s syndrome | hsa-miR-30b-5p↑, miR-150-5p↑, miR-155-5p↑, miR-223-5↑& miR-342-3p↑ | BAFF & B cell proliferation↑ (87,88) |
| Anti-phospholipid syndrome | miR-19b↑ & miR-20a | VEGF & angiogenesis↑ (89) |
| Type 1 diabetes | miR-202-3p↑, miR-326↑ & miR-342↑ | Ongoing autoimmunity (92) |
| miR-34a↓ & miR-146a↓ | Cytokine-mediated b-cell dysfunction (93) | |
| Multiple sclerosis | miR-146↑ & miR-155↑ | TLR-4 activation↑ (99) |
| Myasthenia gravis | miR-21-5p↑ & miR-150-5p↑ | T cell dysfunction & hyperplastic thymus (95) |
| let-7↑ | TLR-7 & T cell activation↑ (95) | |
| Graves’ disease | let-7g-3p↑ & miR-339-5p↑, miR-23b-5p↓ & miR-92a-39↓ | IL-1β↑, TNF-α↑ & Treg↓ (96,97) |
Aberrant miRNA expression profiles in inflammatory rheumatic diseases
Ankylosing spondylitis (AS)
AS is a common and genetically based heterozygous inflammatory rheumatic disease featured by inflammation of the axial and peripheral joints, new bone formation, and spinal ankylosis. High levels of miR-146a-5p, miR-151a-3p, miR-125a-5p and miR-22-3p expression as well as low levels of miR-150-5p, and miR-451a have been shown in AS. Furthermore, miR-146a-5p, miR-125a-5p, miR-151a-3p, miR-22-3p and miR-451a are more likely to be associated with AS than to be associated with psoriatic arthritis. On the other hand, miR-146a-5p, miR-125a-5p and miR-22-3p expression is increased in active versus inactive status of AS. miR-125a-5p, miR-151a-3p, miR-150-5p and miR-451a are relevant to the development of syndesmophytes in AS (99-101).
Psoriasis and psoriatic arthritis (PsA)
Psoriasis is a chronic inflammatory skin disease caused by a complex interplay among the immune system, keratinocytes, susceptibility genes, and environmental triggers. miRNAs may be a possible class of sncRNAs which regulate psoriasis gene expression. Mounting evidence has supported miRNAs as an important triggering etiology in the pathogenesis of psoriasis as well as PsA and other chronic inflammatory conditions. miRNAs including miR-203 and miR-125b have been identified from psoriatic skin, blood, and hair samples and were found associated with non-suppressive effect. On the other hand, miR-146a is associated with psoriasis susceptibility (102).
miR-203 and miR-125b are implicated in hyperproliferative status of psoriasis. A number of authors have suggested that circulating miRNAs from blood samples can become potential biomarkers for diagnosis, monitoring disease activity and foreseeing treatment outcome in psoriasis and allied diseases (102-104).
IBDs
IBD is characterized by a chronic inflammation in the lower gastrointestinal tract. Crohn’s disease and ulcerative colitis are the two main disease entities of IBD. Crohn’s disease may involve the whole digestive tract, beginning from the oral cavity. In contrast, ulcerative colitis is almost confined to the lower gastrointestinal tract. Cao et al. (105) reviewed the role of miRNAs in IBD for diagnosis and correlation with disease activity. Some promising miRNAs including miR-19a, miR-21, miR-31, miR-101, miR-146a and miR-375 have been elaborated for general diagnosis and disease activity monitoring. In addition, a role of miRNAs in IBD-related acne prediction as well as prognosis telling has also been suggested (106).
Coeliac disease (CD)
CD is an autoimmune enteropathy triggered by the interplay between genetic predisposition (HLA-DQ2 or HLA-DQ8) and dietary gluten to induce inflammatory process and to cause bowel mucosa destruction. Dysregulated intestinal miRNA expression such as miR-31-5p, miR-192, miR-194, miR-449a and miR-638 has been reported to correlate with Wnt signaling, cell proliferation and differentiation. Felli et al. (107) suggested that these dysfunctional miRNAs are potentially the disease biomarker of CD.
Table 4 summarizes various ncRNA expression-mediated pathological processes in different inflammatory rheumatic diseases.
Table 4
| Disease | Bio-signature | Pathological Process |
|---|---|---|
| Ankylosing spondylitis | let-7i↑ | Th1 (IFN-γ) immune response↑ (100) |
| miR-21↑, miR-124↑, let-7i↑ & miR-130a↓ | Bone erosion↑, inflammation↑, TNF-α↑, autophagy↑ & TLR-4 down regulation (102) | |
| Psoriasis | miR-21↓, miR-31↑, miR-125a↑ & miR-146a↑ | T cell apoptosis↓ |
| Keratinocyte-immune interaction↑ | ||
| Chronic skin inflammation↑ & | ||
| Epidermal differentiation↑ | ||
| Psoriatic arthritis | miR-146a↑ | IL-1R associated kinase↓ & TRAF-6↓ (104) |
| miR-21-5p↑ | Inflammatory process↑ (105) | |
| Inflammatory bowel disease | miR-126a↑, miR146b↑ & miR-155↑ | NK-kB↑ & pro-inflammatory cytokines↑ (106) |
| miR-214↑ & miR-224↑ | P21 expression↓ & late neoplastic progression↑ (107) | |
| Coeliac disease | miR-31-5p↑, miR-192↑, miR194↑, miR-449a↑ & miR-638↑ | Wnt signal↑, cell proliferation/differentiation↑ & adherent junction pathway↑ (108) |
Conclusions and prospective
miRNAs are evolutionally conserved key players for cellular and developmental process in eukaryotic organism at the post-transcriptional level. However, these miRNA molecules target mRNA in a non-specific manner so that a miRNA can target several mRNAs, or alternatively, several miRNAs may target the same mRNA molecule. By way of microarray detection and bioinformatic analysis, miRNA expression profile may potentially become bio-signatures for etiopathogenesis, general diagnosis, disease activity, therapeutic monitoring, and prognosis. The interpretation of these bio-signatures from high throughput “omics” resulted in somewhat difficult and inconsistency. A useful disease biomarker must primarily be the disease-causing molecule with high sensitivity and specificity in predicting disease risk or monitoring the disease activity. The miRNAs can be obtained from plasma, tissue fluid, specific tissues or immune-related cells. How to direct correlate, but not only associate, miRNAs with disease entity should be considered. It is suggested miRNAs be extracted from a particular tissue (e.g., the kidney) or a particular specimen (e.g., urine or body fluid) and be compared for the cause-effect relationship between them for diagnosis, determining disease activity, therapeutic monitoring and prognosis prediction (70-73). Furthermore, the functional studies of the involved ncRNAs in the targeted tissues are equally important for understanding the pathogenesis and pathological processes of the individual disease entities.
For searching a useful disease biomarker/bio-signature, lncRNAs (>250 bp) are considered more suitable candidates than miRNAs due to their cell lineage-specificity in contrast to the pleotropic properties of miRNAs. Up to the present, only a limit of studies have been reported in autoimmune diseases such as RA (78,79). Recently, a new type of RNA, named circular RNAs (cirRNAs) based on its covalently closed structure, was extensively studied in eukaryotic cells. These cirRNAs have been found lack of terminated 5' caps and 3' tails. These molecules can compete with linear RNAs with regards to tissue specificity by regulating RNA splicing and working as endogenous sponge RNAs to bounce mRNAs (108). Li et al. (109) have demonstrated that plasma cirRNA profile could be used as novel bio-signature for SLE. It is expected that more and more lncRNAs and cirRNAs will be found and be used for comparison with miRNAs in different autoimmune and inflammatory rheumatic diseases in the near future. However, it is more practical at the present time to use miRNA as diagnostic biomarker by taking into account the criteria listed in Table 1.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2018.05.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tsokos GC, Lo MS, Reis PC, et al. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol 2016;12:716-30. [Crossref] [PubMed]
- Noulton VR, Suarez-Fueyo A, Meidan E, et al. Pathogenesis of human systemic lupus erythematosus: a celluar perspective. Trend Mol Med 2017;23:615-35. [Crossref]
- Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus. J Autoimmun 2015;64:125-36. [Crossref] [PubMed]
- Deng Y, Tsao BP. Updates in lupus genetics. Curr Rheumatol Rep 2017;19:68. [Crossref] [PubMed]
- Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr Opin Rheumatol 2014;26:482-92. [Crossref] [PubMed]
- Wu H, Zhao M, Chang C, et al. The real culprit in systemic lupus erythematosus: abnormal epigenetic regulation. Int J Mol Sci 2015;16:11013-33. [Crossref] [PubMed]
- Long H, Yin H, Wang L, et al. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J Autoimmun 2016;74:118-38. [Crossref] [PubMed]
- Invernizzi P, Pasini S, Selmi C, et al. Female predominance and X chromosome defects in autoimmune diseases. J Autoimmun 2009;33:12-6. [Crossref] [PubMed]
- Quintero OL, Amador-Patarroyo MJ, et al. Autoimmune diseases and gender: plausible mechanisms for the female predominance of autoimmunity. J Autoimmun 2012;38:J109-19. [Crossref] [PubMed]
- Hughes GC, Choubey D. Modulation of autoimmune rheumatic diseases by estrogen and progesterone. Nat Rev Rheumatol 2014;10:740-51. [Crossref] [PubMed]
- Nie J, Li YY, Zheng SG, et al. FOXP3(+) Treg Cells and Gender Bias in Autoimmune Diseases. Front Immunol 2015;6:493. [Crossref] [PubMed]
- Vojdani A. A potential link between environmental triggers and autoimmunity. Autoimmune Dis 2014;2014:437231.
- Gulati G, Brunner HI. Environmental triggers in systemic lupus erythematosus. Semin Arthritis Rheum 2018;47:710-17. [Crossref] [PubMed]
- Kim D, Yoo SA, Kim WU. Gut microbiota in autoimmunity: potential for clinical applications. Arch Pharm Res 2016;39:1565-76. [Crossref] [PubMed]
- de Oliveira GL, Leite AZ, Higuchi BS, et al. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 2017;152:1-12. [Crossref] [PubMed]
- Felix KM, Tahsin S, Wu HJ. Host-microbiota interplay in mediating immune disorders. Ann N Y Acad Sci 2018;1417:57-70. [Crossref] [PubMed]
- Rosser EC, Mauri C. A clinical update on the significance of the gut microbiota in systemic autoimmunity. J Autoimmun 2016;74:85-93. [Crossref] [PubMed]
- Rodríguez-Carrio J, López P, Sánchez B, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front Immunol 2017;8:23. [Crossref] [PubMed]
- Roberts-Thomson PJ, Walker JG. Stochastic processes in the etiopathogenesis of scleroderma. Intern Med J 2012;42:235-42. [Crossref] [PubMed]
- Tuohy VK, Kinkel RP. Epitope spreading: a mechanism for progression of autoimmune diseases. Arch Immunol Ther Exp (Warsz) 2000;48:347-51. [PubMed]
- Cornaby C, Gibbons L, Mayhew V, et al. B cell epitope spreading: mechanisms and contribution to autoimmune diseases. Immunol Lett 2015;163:56-68. [Crossref] [PubMed]
- Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modification and DNA methylation in gene silencing. Mutat Res 2008;659:40-8. [Crossref] [PubMed]
- Dieker J, Muller S. Epigenetic histone code and autoimmunity. Clin Rev Allergy Immunol 2010;39:78-84. [Crossref] [PubMed]
- Zhao M, Wang Z, Yung S, et al. Epigenetic dynamics in immunity and autoimmunity. Int J Biochem Cell Biol 2015;67:65-74. [Crossref] [PubMed]
- Zhang Y, Zhao M, Sawalha AH, et al. Impaired DNA methylation and its mechanism in CD4(+) T cells of systemic lupus erythematosus. J Autoimmun 2013;41:92-9. [Crossref] [PubMed]
- Miao CG, Yang JT, Yang YY, et al. Critical role of DNA methylation in the pathogenesis of systemic lupus erythematosus: new advances and future challenges. Lupus 2014;23:730-42. [Crossref] [PubMed]
- Wu H, Zhao M, Tan L, et al. The key culprit in the pathogenesis of systemic lupus erythematosus: aberrant DNA methylation. Autoimmun Rev 2016;15:684-9. [Crossref] [PubMed]
- Hawtree S, Muthana M, Davie JR. The role of histone deacetylase in rheumatoid arthritis fibroblast-like synoviocytes. Biochem Soc Trans 2013;41:783-8. [Crossref] [PubMed]
- Wang Z, Yin H, Lau CS, et al. Histone posttranslational modifications of CD4+T cell in autoimmune diseases. Int J Mol Sci 2016;17:1547. [Crossref] [PubMed]
- Araki Y, Mimura T. The histone modification code in the pathogenesis of autoimmune diseases. Mediators Inflamm 2017;2017:2608605.
- Montagner S, Deho L. Moticelli. microRNAs in hematopoietic development. BMC Immunol. 2014;15:14. [Crossref] [PubMed]
- Catalanotto C, Cogoni C, Zardo G. microRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci 2016;17: [Crossref] [PubMed]
- Mehta A, Baltimore D. microRNAs as regulatory elements in immune system logic. Nat Rev Immunol 2016;16:279-94. [Crossref] [PubMed]
- Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long non-coding RNAs. Nat Immunol 2017;18:962-72. [Crossref] [PubMed]
- Tang Y, Zhou T, Yu X, et al. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol 2017;13:657-69. [Crossref] [PubMed]
- Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J 2017;474:4219-51. [Crossref] [PubMed]
- Korte EA, Gaffney PM, Powell DW. Contributions of mass spectrometry-based proteomics to defining cellular mechanisms and diagnostic markers for systemic lupus erythematosus. Arthritis Res Ther 2012;14:204. [PubMed]
- Kazemipour N, Qazizadeh H, Sepehrimanesh M, et al. Biomarkers identified from serum proteomic analysis for the differential diagnosis of systemic lupus erythematosus. Lupus 2015;24:582-7. [Crossref] [PubMed]
- Nicolaou O, Kousios A, Hadjisavvas A, et al. Biomarkers of systemic lupus erythematosus identified using mass spectrometry-based proteomics: a systemic review. J Cell Mol Med 2017;21:993-1012. [Crossref] [PubMed]
- Yan B, Huang J, Zhang C, et al. Serum metabolomic profiling in patients with systemic lupus erythematosus by GC/MS. Mod Rheumatol 2016;26:914-22. [Crossref] [PubMed]
- Yan B, Huang J, Dong F, et al. Urinary metabolomic study of systemic lupus erythematosus based on gas chromatography/mass spectrometry. Biomed Chromatogr 2016;30:1877-81. [Crossref] [PubMed]
- Rhoads JP, Major AS, Rathmell JC. Fine tuning of immunometabolism for the treatment of rheumatic diseases. Nat Rev Rheumatol 2017;13:313-20. [Crossref] [PubMed]
- Jung JY, Bae CB, Suh CH. Promising biomarkers for systemic lupus erythematosus. Expert Opin Med Diagn 2013;7:601-13. [Crossref] [PubMed]
- Rai G, Rai R, Saeidian AH, et al. Microarray to deep sequencing: transcriptome and miRNA profiling to elucidate molecular pathways in systemic lupus erythematosus. Immunol Res 2016;64:14-24. [Crossref] [PubMed]
- Tang D, Chen Y, He H, et al. Integrated analysis of mRNA, microRNA and protein in systemic lupus erythematosus-specific induced pluripotent stem cells from urine. BMC Genomics 2016;17:488. [Crossref] [PubMed]
- Hudson M, Bernatsky S, Colmegna I, et al. Novel insights into systemic autoimmune rheumatic diseases using shared molecular signatures and an integrative analysis. Epigenetics 2017;12:433-40. [Crossref] [PubMed]
- McDermott JE, Wang J, Mitchell H, et al. Challenges in biomarker discovery: combinding expert insights with statistical analysis of complex omics data. Expert Opin Med Diagn 2013;7:37-51. [Crossref] [PubMed]
- Byrnes SA, Weigl BH. Selecting analytical biomarkers for diagnostic applications: a first principles approach. Expert Rev Mol Diagn 2018;18:19-26. [Crossref] [PubMed]
- Khanduja JS, Calvo IA, Joh RI, et al. Nuclear non-coding RNAs and genome stability. Mol Cell 2016;63:7-20. [Crossref] [PubMed]
- Sana J, Faltejskova P, Svoboda M. Novel classes of non-coding RNAs and cancer. J Transl Med 2012;10:103. [Crossref] [PubMed]
- Nagy Z, Igaz P. Introduction to microRNAs: biogenesis, action, relevance to tissue microRNAs in disease pathogenesis, diagnosis and therapy-the concept of circulating microRNAs. EXS 2015;106:3-30. [Crossref] [PubMed]
- Bartel DP. MicroRNA target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Bartel DP. Metazoan microRNAs. Cell 2018;173:20-51. [Crossref] [PubMed]
- Dooley J, Linterman MA, Liston A. microRNA regulation of T-cell development. Immunol Rev 2013;253:53-64. [Crossref] [PubMed]
- Sullivan RP, Leong JW, Fehniger TA. MicroRNA regulation of natural killer cells. Front Immunol 2013;4:44. [Crossref] [PubMed]
- Singh RP, Massachi I, Manickavel S, et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev 2013;12:1160-5. [Crossref] [PubMed]
- Chen JQ, Papp G, Szodoray P, et al. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev 2016;15:1171-80. [Crossref] [PubMed]
- Esmailzadeh S, Mansoori B, Mohammadi A, et al. Regulatory roles of micro-RNAs in T cell autoimmunity. Immunol Invest 2017;46:864-79. [Crossref] [PubMed]
- Stagakis E, Bertsias G, Verginis P, et al. Identification of novel microRNA signatures linked to human lupus disease activity and pathogenesis: miR-21 regulates T cell responses through regulation of PDCD4 expression. Ann Rheum Dis 2011;70:1496-506. [Crossref] [PubMed]
- Luo X, Yang W, Ye DQ, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet 2011;7:e1002128 [Crossref] [PubMed]
- Qin H, Zhu X, Liang J, et al. microRNA-29b contributes to DNA hypomethylation of CD4+T cells in systemic lupus erythematosus by indirectly targeting DNA methyl-transferase 1. J Dermatol Sci 2013;69:61-7. [Crossref] [PubMed]
- Lu MC, Lai NS, Chen HC, et al. Decreased microRNA(miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin Exp Immunol 2013;171:91-9. [Crossref] [PubMed]
- Miao CG, Yang YY, He X, et al. The emerging role of microRNAs in the pathogenesis of systemic lupus erythematosus. Cell Signal 2013;25:1828-36. [Crossref] [PubMed]
- Yan S, Yim LY, Lu L, et al. MicroRNA regulation in systemic lupus erythematosus pathogenesis. Immune Netw 2014;14:138-48. [Crossref] [PubMed]
- Lu MC, Yu CL, Chen HC, et al. Aberrant T cell expression of Ca2+ influx-regulated miRNAs in patients with systemic lupus erythematosus promotes lupus pathogenesis. Rheumatology (Oxford) 2015;54:343-8. [Crossref] [PubMed]
- Amr KS, Bayoumi FS, Elgengehy FT, et al. The role of microRNA-31 and microRNA-21 as regulatory biomarkers in the activation of T lymphocytes of Egyptian lupus patients. Rheumatol Int 2016;36:1617-25. [Crossref] [PubMed]
- Li LJ, Zhao W, Tao SS, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol 2017;319:17-27. [Crossref] [PubMed]
- Lai NS, Koo M, Yu CL, et al. Immunopathogenesis of systemic lupus erythematosus and rheumatoid arthritis: the role of aberrant expression of non-coding RNAs in T cells. Clin Exp Immunol 2017;187:327-36. [Crossref] [PubMed]
- Stypińska B, Paradowska-Gorycka A. Cytokines and microRNAs as candidate biomarkers for systemic lupus erythematosus. Int J Mol Sci 2015;16:24194-218. [Crossref] [PubMed]
- Abulaban KM, Fall N, Nurna R, et al. Relationship of cell-free urine microRNA with lupus nephritis. Pediatr Rheumatol Online J 2016;14:4-10. [Crossref] [PubMed]
- Hsieh SC, Tsai CY, Yu CL. Potential serum and urine biomarkers in patients with lupus nephritis and the unsolved problems. Open Access Rheumatol 2016;8:81-91. [Crossref] [PubMed]
- Cardenas-Gonzalez M, Srivastava A, Pavkovic M, et al. Identification, confirmation, and replication of novel urinary microRNA biomarkers in lupus nephritis and diabetic nephropathy. Clin Chem 2017;63:1515-26. [Crossref] [PubMed]
- Tsai CY, Lu MC, Yu CL. Can urinary exosomal microRNA detection become a diagnostic and prognostic gold standard for patients with lupus nephritis and diabetic nephropathy? J Lab Precision Med 2017;2:91. [Crossref]
- Fu L, Jin L, Yan L, et al. Comprehensive review of genetic association studies and meta-analysis on miRNA polymorphisms and rheumatoid arthritis and systemic lupus erythematosus. Hum Immunol 2016;77:1-6. [Crossref] [PubMed]
- Ammari M, Jorgensen C, Apparailly F. Impact of microRNAs on the understanding and treatment of rheumatoid arthritis. Curr Opin Rheumatol 2013;25:225-33. [Crossref] [PubMed]
- Philippe L, Alsaleh G, Bahram S, et al. The miR-17~92 cluster: a key player in the control of inflammation during rheumatoid arthritis. Front Immunol 2013;4:70. [Crossref] [PubMed]
- Lu MC, Yu CL, Chen HC, et al. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol 2014;177:641-51. [Crossref] [PubMed]
- Lu MC, Yu HC, Yu CL, et al. Increased expression of long non-coding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol Res 2016;64:576-83. [Crossref] [PubMed]
- Sharma AR, Sharma G, Lee SS, et al. miRNA-regulated key components of cytokine signaling pathways and inflammation in rheumatoid arthritis. Med Res Rev 2016;36:425-39. [Crossref] [PubMed]
- Lai NS, Yu HC, Tung CH, et al. The role of aberrant expression of T cell miRNAs affected by TNF-α in the immunopathogenesis of rheumatoid arthritis. Arthritis Res Ther 2017;19:261. [Crossref] [PubMed]
- Peng JS, Chen SY, Wu CL, et al. Amelioration of experimental autoimmune arthritis through targeting of fibroblasts by intra-articular delivery of microRNAs 140-3p and 140-5p. Arthritis Rheumatol 2016;68:370-81. [Crossref] [PubMed]
- Tavasolian F, Abdollahi E, Rezaei R, et al. altered expression of microRNAs in rheumatoid arthritis. J Cell Biochem 2018;119:478-87. [Crossref] [PubMed]
- Shi DL, Shi GR, Xie J, et al. MicroRNA-27a inhibits cell migration and invasion of fibroblast-like synoviocytes by targeting follistatin-like protein 1 in rheumatoid arthritis. Mol Cells 2016;39:611-8. [Crossref] [PubMed]
- Sujitha S, Rasool M. MicroRNAs and bioactive compounds on TLR/MAPK signaling in rheumatoid arthritis. Clin Chim Acta 2017;473:106-15. [Crossref] [PubMed]
- Lai NS, Yu HC, Yu CL, et al. Anti-citrullinated protein antibodies suppress let-7a expression in monocytes from patients with rheumatoid arthritis and facilitate the inflammatory response in rheumatoid arthritis. Immunobiology 2015;220:1351-8. [Crossref] [PubMed]
- Alevizos I, Illei GG. MicroRNAs in Sjögren’s syndrome as a prototypic autoimmune disease. Autoimmun Rev 2010;9:618-21. [Crossref] [PubMed]
- Chen JQ, Papp G, Poliska S, et al. MicroRNA expression profiles identity disease-specific alterations in systemic lupus erythematosus and primary Sjögren’s syndrome. PLoS One 2017;12:e0174585 [Crossref] [PubMed]
- Wang-Renault SF, Bovdaoud S, Noiturne G, et al. Deregulation of microRNA expression in purified T and B lymphocytes from patients with primary Sjögren’s syndrome. Ann Rheum Dis 2018;77:133-40. [Crossref] [PubMed]
- Muhammad Shazwan S, Muhammad Aliff M, Asral Wirda AA, et al. MicroRNA expression in antiphospholipid syndrome: a systemic review and microRNA target genes analysis. Malays J Pathol 2016;38:273-83. [PubMed]
- Perez-Sánchez C, Arias-de la Rosa I, Aguirre MÁ, et al. Circulating microRNAs as biomarkers of disease and typification of the atherothrombotic status in antiphospholipid syndrome. Haematologica 2018;103:908-18. [Crossref] [PubMed]
- Steen SO, Iversen LV, Carlsen AL, et al. The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J Rheumatol 2015;42:214-21. [Crossref] [PubMed]
- Isaacs SR, Wang J, Kim KW, et al. MicroRNAs in type 1 diabetes: complex inter-regulation of the immune system, β cell function and viral infections. Curr Diab Rep 2016;16:133. [Crossref] [PubMed]
- Zheng Y, Wang Z, Zhou Z. miRNAs: novel regulators of autoimmune-mediated pancreatic β-cell destruction in type 1 diabetes. Cell Mol Immunol 2017;14:488-96. [Crossref] [PubMed]
- Assmann TS, Recamonde-Mendoza M, De Souza BM, et al. MicroRNA expression profiles and type 1 diabetes mellitus: systemic review and bioinformatic analysis Endocr Connect 2017;6:773-90. [Crossref] [PubMed]
- Punga AR, Punga T. Circulating microRNAs as potential biomarkers in myasthenia gravis patients. Ann NY Acad Sci 2018;1412:33-40. [Crossref] [PubMed]
- Wang Z, Fan X, Zhang R, et al. Integrative analysis of mRNA and miRNA array data reveals the suppression of retinoic acid pathway in regulatory T cells of Graves’ disease. J Clin Endocrinol Metab. 2014;99:E2620-7. [Crossref] [PubMed]
- Hiratsuka I, Yamada H, Munetsuna E, et al. Circulating microRNAs in Graves’ disease in relation to clinical activity. Thyroid 2016;26:1431-40. [Crossref] [PubMed]
- Kacperska MJ, Walenczak J, Tomasik B. Plasmatic microRNA as potential biomarkers of multiple sclerosis. Adv Clin Exp Med 2016;25:775-9. [Crossref] [PubMed]
- Lai NS, Yu HC, Chen HC, et al. Aberrant expression of microRNAs in T cells from patients with ankylosing spondylitis contributes to the immunopathogenesis. Clin Exp Immunol 2013;173:47-57. [Crossref] [PubMed]
- Perez-Sanchez C, Font-Ugalde P, Ruiz-Limon P, et al. Circulating microRNAs as potential biomarkers of diseases activity and structural damage in ankylosing spondylitis patients. Hum Mol Genet 2018;27:875-90. [Crossref] [PubMed]
- Mohammadi H, Hemmatzadeh M, Babaie F, et al. MicroRNA implications in the etiopathogenesis of ankylosing spondylitis. J Cell Physiol 2018;233:5564-73. [Crossref] [PubMed]
- Sonkoly E. The expending microRNA would in psoriasis. Exp Dermatol 2017;26:375-6. [Crossref] [PubMed]
- Chatzikyriakidou A, Voulgari PV, Georgiou I, et al. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol 2010;71:382-5. [Crossref] [PubMed]
- Ciancio G, Ferracin M, Saccenti E, et al. Characterization of peripheral mononuclear cell microRNA in early onset psoriatic arthritis. Clin Exp Rheumatol 2017;35:113-121. [PubMed]
- Cao B, Zhou X, Ma J, et al. Role of miRNAs in inflammatory bowel disease. Dig Dis Sci 2017;62:1426-38. [Crossref] [PubMed]
- Olaru AV, Yamanaka S, Vazquez C, et al. MicroRNA-224 negatively regulates p21 expression during late neoplastic progression in inflammatory bowel disease. Inflamm Bowel Dis 2013;19:471-80. [Crossref] [PubMed]
- Felli C, Baldassarre A, Masotti A. Intestinal and circulating mcroRNA in coeliac disease. Int J Mol Sci 2017;18:1907. [Crossref] [PubMed]
- Liu J, Liu T, Wang X, et al. Circles reshaping the RNA world: from waste to treasure. Mol Cancer 2017;16:58. [Crossref] [PubMed]
- Li H, Li K, Lai W, et al. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta 2018;480:17-25. [Crossref] [PubMed]
Cite this article as: Tsai CY, Hsieh SC, Lu MC, Yu CL. Aberrant non-coding RNA expression profiles as biomarker/bio-signature in autoimmune and inflammatory rheumatic diseases. J Lab Precis Med 2018;3:51.