Glioblastoma biomarkers: finding a needle in a haystack
Introduction
Glioblastoma, also known as glioblastoma multiforme, is one of the most aggressive and lethal brain cancers. Although this malignancy is considered relative rare (the overall prevalence is approximately 3–10:100,000 persons, accounting for ~2.5% of all cancer deaths), life expectancy is extremely poor, usually comprised between 3–5% at five years, with the vast majority of patients dying within 15 months from initial diagnosis (1). The diagnostic approach of patients with glioblastoma is essentially based on imaging techniques, namely magnetic resonance imaging (MRI) using either traditional or advanced technologies or, less frequently, computed tomography (CT), along with (stereotactic needle) tissue biopsy or tumor resection, followed by pathologic analysis (1).
Although many different therapeutic options have been attempted over the past decades (i.e., surgical ablation, radiation, chemotherapy, immunotherapy, anti-angiogenic drugs and epigenetic modulators) (2-4), most of these have been largely unsuccessful to substantially reverse disease progression, so that the prognosis of this type of cancer remains dramatically poor. Therefore, an early diagnosis of glioblastoma seems now the most effective strategy for improving the outcome or, at least, for prolonging progression-free survival of affected patients.
Glioblastoma biomarkers
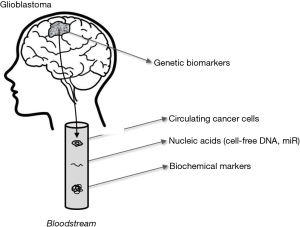
Although it is now unquestionable that laboratory diagnostics strongly contributes to the screening and diagnosis of many human conditions, laboratory tests in cancer diagnostics are prevalently useful for therapeutic management and early detection of recurrence. Glioblastoma makes no exception to this rule, but represents an even harder challenge due to many aspects including cancer cell heterogeneity and the fact that the blood-brain barrier further reduces the likelihood that some potentially useful biomarkers, actively released by cancer cells, will early reach the bloodstream where they can then be efficiently assessed. As for many other types of cancers, the putative biomarkers for early detection/diagnosis/monitoring of glioblastoma can be divided in four main categories, encompassing genetic, epigenetic and biochemical biomarkers, as well as circulating tumor cells or DNA (Figure 1, Table 1).
Table 1
| Genetic markers |
| Receptor tyrosine kinase (RTK) |
| Epidermal growth factor receptor (EGFR) |
| Platelet-derived growth factor receptor alpha (PDGFRA) |
| Basic fibroblast growth factor receptor 1 (FGFR-1) |
| Insulin-like growth factor receptor (IGFR-1) |
| TP53 tumor suppressor gene |
| Retinoblastoma protein (RB1) protein pathway |
| Cyclin dependent kinase 2a/p16 (CDKN2A/p16) |
| Cyclin-dependent kinase 4 (CDK4) |
| Isocitrate dehydrogenase 1 (IDH1) and 2 (IDH2) |
| Phosphatase and tensin homolog (PTEN) |
| Loss of heterozygosity (LOH) on chromosome 10q |
| 1p/19q co-deletion |
| Interleukin-13 (IL-13) receptor α1 and α2 |
| Alpha-thalassemia/mental retardation X-linked gene (ATRX) |
| Telomerase reverse transcriptase-encoding gene (TERT) |
| Epigenetic biomarkers |
| Methylguanine-DNA-methyltransferase (MGMT) promoter methylation |
| Micro RNAs |
| miR-21 |
| miR-10b |
| miR-454-3p |
| miR-222 |
| miR-124-3p |
| Biochemical biomarkers |
| Immunosuppressive acidic protein |
| Alpha-1 acidic glycoprotein, |
| Alpha-1 antitrypsin |
| Fibronectin |
| Endothelial cell-derived thrombomodulin-1 |
| Vascular endothelial growth factor (VEGF) |
| Placental growth factor (PlGF) |
| Platelet-derived growth factor (PDGF) |
| Specific neural proteins |
| Glial fibrillary acidic protein (GFAP) |
| Brain-derived neurotrophic factor (BDNF) |
| Protein S100 B (S100B) |
| Neural cell adhesion molecule (NCAM) |
| 2-hydroxyglutarate (2-HG) |
| Chitinase-3-like protein 1 (CHI3L1), |
| Interleukin-2 (IL-2) |
| Transforming growth factor-β (TGF-β) |
| Tumor necrosis factor-α (TNF-α) |
| Complement component C9 (C9) |
| C-reactive protein (CRP) |
| Leucine-rich alpha-2-glycoprotein (LRG1) |
| Gelsolin |
| Apolipoprotein A-IV (APOA4) |
| Ig alpha-1 chain C region (IGHA1) |
| Matrix metalloproteinases (MMPs) |
| MMP-2 |
| MMP-9 |
| MMP-10 |
| Tissue inhibitors of metalloproteinases (TIMPs) |
| TIMP-1 |
| Traditional cancer biomarkers |
| Carcinoembryonal antigen (CEA) |
| Human chorionic gonadotropin (hCG) |
| Alpha-fetoprotein (AFP) |
Regarding the former class, the most studied genetic alterations include mutations or amplifications of genes encoding for growth proteins and cell signaling, namely those belonging to the receptor tyrosine kinase (RTK) signaling [i.e., epidermal growth factor receptor (EGFR), platelet-derived growth factor receptor alpha (PDGFRA), basic fibroblast growth factor receptor 1 (FGFR-1) and insulin-like growth factor receptor (IGFR-1)], TP53 tumor suppressor gene, retinoblastoma protein (RB1) protein pathway [especially those of cyclin dependent kinase 2a/p16 (CDKN2A/p16) and cyclin-dependent kinase 4 (CDK4)], isocitrate dehydrogenase 1 (IDH1) and 2 (IDH2), phosphatase and tensin homolog (PTEN), loss of heterozygosity (LOH) on chromosome 10q, 1p/19q co-deletion, interleukin-13 (IL-13) receptor α1 and α2, alpha-thalassemia/mental retardation X-linked gene (ATRX) and telomerase reverse transcriptase-encoding gene (TERT) (5-8) (Table 1). Albeit the study of these molecular abnormalities is indeed promising not only for glioblastoma diagnostics, but also for developing targeted (personalized) therapies against the involved genetic mutations, their clinical usefulness has been limited so far by the fact that they can be mainly used as prognostic or predictive biomarkers in cancer tissues, whilst their use as early diagnostic biomarkers (i.e., liquid biopsy) has been by far less explored.
The most studied glioblastoma epigenetic biomarkers include the methylguanine-DNA-methyltransferase (MGMT) promoter methylation status, some micro RNAs (miRs) such as miR-21, miR-10b, miR-454-3p, miR-222 and miR-124-3p, among others (5,9,10) (Table 1). Notably, a recent epigenetic study carried out in our laboratory allowed to identify a serum exosome signature (i.e., miR-21, miR-222 and miR-124-3p) from patients with glioma, which was characterized by 87% diagnostic accuracy for discriminating cancer patients from healthy controls, as well 83% diagnostic accuracy for discriminating patients with low-grade from those with high-grade cancer (10). This study clearly highlights that the investigation of some exosome-associated miRNAs should strengthen the diagnostic specificity of these biomarkers. Unlike somatic mutations, miRNA have the important advantage that can now be easily, rapidly and economically measured in blood samples, thus representing a less invasive and cumbersome approach compared to other conventional strategies.
Regarding circulating biochemical biomarkers, several studies showed increased concentrations of several proteins, cytokine and hormones, including immunosuppressive acidic protein, alpha-1 acidic glycoprotein, alpha-1 antitrypsin, fibronectin, endothelial cell-derived thrombomodulin-1, vascular endothelial growth factor (VEGF), placental growth factor (PlGF), platelet-derived growth factor (PDGF), specific neuronal protein [i.e., glial fibrillary acidic protein (GFAP), brain-derived neurotrophic factor (BDNF), protein S100 B (S100B), neural cell adhesion molecule (NCAM)], of 2-hydroxyglutarate (2-HG), chitinase-3-like protein 1 (CHI3L1), interleukin-2 (IL-2), transforming growth factor-β (TGF-β), tumor necrosis factor-α (TNF-α), matrix metalloproteinases (MMPs) or tissue inhibitors of metalloproteinases (TIMPs) such as MMP-2, MMP-9, MMP-10 and TIMP-1, as well as that of other more traditional cancer biomarkers (Table 1) (9). Unfortunately, the diagnostic efficiency of all these biomarkers remains relatively low in glioblastoma diagnostics, especially those which are reportedly not tumor-specific (i.e., VEGF, cytokines, MMPs, traditional cancer biomarkers). Interestingly, in a recent study based on a proteomic approach, eight putative biomarkers for glioblastoma could be identified. Six of these proteins, namely over-expression of complement component C9 (C9), C-reactive protein (CRP), leucine-rich alpha-2-glycoprotein (LRG1) and under-expression of gelsolin, apolipoprotein A-IV (APOA4) and Ig alpha-1 chain C region (IGHA1), yielded a diagnostic efficiency higher than 80%, whilst the concentration of three of these (i.e., C9, CRP and LRG1) was also significantly correlated with tumor size (11).
The assessment of circulating tumor cells is indeed a valuable perspective for diagnostics of many types of cancers. Albeit their early detection in patients with glioblastoma would be plagued by the same drawbacks as those of other biomarkers (e.g., the presence of the brain-blood barriers, which retards the entrance of tumor material into the bloodstream), the use of fluorescence immunocytochemistry has recently allowed to distinguishing advanced (i.e., metastatic) from localized cancer. Some ongoing studies, using fluorescent probes recognizing specific glioma cells markers (i.e., GFAP, EGFR, Sox2, Tubulin beta-3, A2B5 and c-Met), are now investigating the potential clinical usefulness of this approach for early detection of metastatic cancer (9). Another interesting option is the possibility of detecting cell-free circulating tumour DNA. Although is has not been validated for clinical use so far, promising evidence has been provided that circulating DNA bearing suggestive genetic aberrations (e.g., those involving IDH1, MGMT, EGFR, etc.) may be seen as a valuable perspective for identifying primary aggressive or invasive tumors and their metastases (12), since a consistent number of primary patients with glioblastoma may display detectable cell-free circulating tumor DNA even at an early stage (13).
Conclusions and future perspectives
Despite being currently considered a relatively rare cancer, glioblastoma has a considerable clinical economic and social impact due to the limited success that has been achieved so far for its prevention and treatment. Most of the current drawbacks in early diagnostics are attributable to cancer cells heterogeneity and cerebral localization of the tumor (i.e., unfeasible access for easy and frequent sampling, delayed appearance of tumor material in the bloodstream), which both contribute to diminish the potential diagnostic options (12), and make the search for candidate biomarkers more or less like finding a needle in a haystack. However, interesting evidence emerged from recently published studies attests that the assessment of some epigenetic biomarkers (namely MGMT promoter methylation status and combined measurement of miR-21, miR-222 and miR-124-3p), proteomic analysis and identification of cell-free circulating tumor DNA may pave the way to a paradigm shift in glioblastoma diagnostics. It is also conceivable that combining these different strategies, an approach which has not been validated so far, may yield better diagnostic performance that using one single approach alone. Albeit we would all agree that the use of cerebrospinal fluid (CSF) for biomarkers assessment is invasive and carries the risk of important side effects or complications, it is undeniable that the identification of glioblastoma signatures in CSF, either molecular or biochemical (14-16), will perhaps enable to identify localized cancers much earlier than the appearance and detectability of these biomarkers in the bloodstream. Further studies will hence be needed to explore the value of using these promising strategies in the future, as well as their potential impact on personalized treatment of glioblastoma.
Acknowledgments
This work was supported by generous donations from Giovanni Celeghin Foundation ONLUS, Brain Research Foundation Verona (VBRF), and “Fondo Ricerca Indipendente” (FRI), University Hospital of Verona, Italy.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2018.06.04). Giuseppe Lippi serves as the unpaid Editor-in-Chief of Journal of Laboratory and Precision Medicine from November 2016 to October 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hanif F, Muzaffar K, Perveen K, et al. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac J Cancer Prev 2017;18:3-9. [PubMed]
- Davis ME. Glioblastoma: Overview of Disease and Treatment. Clin J Oncol Nurs 2016;20:S2-8. [Crossref] [PubMed]
- Winkler F, Osswald M, Wick W. Anti-Angiogenics: Their Role in the Treatment of Glioblastoma. Oncol Res Treat 2018;41:181-6. [Crossref] [PubMed]
- Lee DH, Ryu HW, Won HR, et al. Advances in epigenetic glioblastoma therapy. Oncotarget 2017;8:18577-89. [PubMed]
- Szopa W, Burley TA, Kramer-Marek G, et al. Diagnostic and Therapeutic Biomarkers in Glioblastoma: Current Status and Future Perspectives. Biomed Res Int 2017;2017:8013575 [Crossref] [PubMed]
- Sasmita AO, Wong YP, Ling APK. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol 2018;14:40-51. [Crossref] [PubMed]
- Han J, Puri RK. Analysis of the cancer genome atlas (TCGA) database identifies an inverse relationship between interleukin-13 receptor α1 and α2 gene expression and poor prognosis and drug resistance in subjects with glioblastoma multiforme. J Neurooncol 2018;136:463-74. [Crossref] [PubMed]
- Hochberg FH, Atai NA, Gonda D, et al. Glioma diagnostics and biomarkers: an ongoing challenge in the field of medicine and science. Expert Rev Mol Diagn 2014;14:439-52. [Crossref] [PubMed]
- Best MG, Sol N, Zijl S, et al. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol 2015;129:849-65. [Crossref] [PubMed]
- Santangelo A, Imbrucè P, Gardenghi B, et al. A microRNA signature from serum exosomes of patients with glioma as complementary diagnostic biomarker. J Neurooncol 2018;136:51-62. [Crossref] [PubMed]
- Miyauchi E, Furuta T, Ohtsuki S, et al. Identification of blood biomarkers in glioblastoma by SWATH mass spectrometry and quantitative targeted absolute proteomics. PLoS One 2018;13:e0193799 [Crossref] [PubMed]
- Kros JM, Mustafa DM, Dekker LJ, et al. Circulating glioma biomarkers. Neuro Oncol 2015;17:343-60. [PubMed]
- Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol 2015;26:1994-9. [Crossref] [PubMed]
- Akers JC, Hua W, Li H, et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 2017;8:68769-79. [Crossref] [PubMed]
- Shen F, Zhang Y, Yao Y, et al. Proteomic analysis of cerebrospinal fluid: toward the identification of biomarkers for gliomas. Neurosurg Rev 2014;37:367-80. [Crossref] [PubMed]
- De Mattos-Arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015;6:8839. [Crossref] [PubMed]
Cite this article as: Lippi G, Dechecchi MC. Glioblastoma biomarkers: finding a needle in a haystack. J Lab Precis Med 2018;3:59.