
Comparison of different immunoassays for γH2AX quantification
Introduction
Cellular biomolecules are constantly harmed by various endogenous and exogenous substances. In contrast to damaged proteins and lipids undergoing general recycling processes based on degradation and synthesis, altered DNA is repaired by different, lesion specific repair mechanisms throughout lifetime instead (1). One of the most severe types of DNA lesions are DNA double-strand breaks (DSB). After DSB formation, rapid phosphorylation of the core histone variant H2AX at serine 139 is induced in adjacent chromatin, leading to the formation of a γH2AX focus (2). Initially, γH2AX was detected by two-dimensional gel electrophoresis, but generation of specific antibodies enabled the development of more sensitive γH2AX immunoassays (2-4). Thereby, γH2AX has been established as one of the most sensitive biomarkers for DNA DSB detection offering multiple fields of application (5-8). Apart from basic research, γH2AX has the potential to be used as diagnostic biomarker in clinics, e.g., for monitoring of radio- or chemotherapy, for biodosimetry, for drug development and to study the impact of environmental toxins or the process of aging (5-7).
Within the last two decades, innumerous reports have been published applying γH2AX immunoassays, including immunocytochemical staining followed by microscopic or flow cytometric analysis, immunoblotting and enzyme-linked immunosorbent assay (ELISA). The most common approach is microscopic quantification of γH2AX foci. Here, single cells as well as single foci can be analyzed. In contrast, flow cytometry only allows overall γH2AX fluorescence intensity measurement of individual cells, but it cannot be differentiated whether the signal originated from one bright focus, many small foci or background intensity. Further, immunoblotting or ELISA only enable a general assessment of total γH2AX protein level of the whole sample, but e.g., apoptotic cells showing a pan-nuclear γH2AX expression cannot be distinguished from viable cells and may thereby influence γH2AX quantification (2).
Fluorescent microscopic foci quantification has been described as one of the most sensitive methods for γH2AX assessment, enabling detection of even one single focus (2,9-11). The aim of the current study was to verify this statement by performing automated fluorescent microscopy, as described in detail elsewhere (12-14), and to compare obtained limits of detection (LoD) with different approaches. Therefore, human peripheral blood mononuclear cells (PBMCs) were exposed to different levels of the DNA DSB-inducing cytostatic drug etoposide for one hour. Afterwards, γH2AX levels were quantified by automated microscopy, flow cytometry and immunoblotting. Here we discuss results of the γH2AX assay comparison as well as assay specific advantages and disadvantages.
Methods
Isolation and treatment of human PBMCs
Heparinized blood was obtained from healthy blood donors and PBMCs were isolated by density gradient centrifugation using Biocoll separating solution (Biochrom, Berlin, Germany). Afterwards, PBMCs were washed and suspended to a final density of 1×106 PBMCs per ml in Roswell Park Memorial Institute (RPMI) 1640 cell culture medium (Biochrom) containing 10% fetal bovine serum (Pan Biotech, Aidenbach, Germany), 100 U/mL penicillin and 100 µg/mL streptomycin (both Life Technologies GmbH, Darmstadt, Germany). For induction of DNA DSBs cells were transferred into 24-well cell culture plates and exposed to etoposide at a final concentration of either 0, 0.05, 0.1, 0.25, 0.5, 1, 2.5, 5, 10, 25, 50, 100 or 250 µM for 1 h at 37 °C with 7% CO2.
γH2AX analysis using automated fluorescent microscopy
After etoposide exposure, cells were harvested, washed in phosphate-buffered saline (PBS) and transferred onto silanized glass slides. Subsequently, samples were fixed for 15 min in formaldehyde, permeabilized in 0.1% Triton X-100 and blocked in PBS containing 1% bovine serum albumin (BSA). For γH2AX detection cells, were incubated at room temperature with an anti-phosphohistone H2AX mouse monoclonal IgG primary antibody (Millipore, Schwalbach, Germany; dilution 1:2,000) for 1 h, washed in blocking buffer and additionally stained for 1 h in the dark with a polyclonal goat anti-mouse IgG antibody conjugated to Alexa-Fluor-488 (Life Technologies, Darmstadt, Germany; dilution 1:2,000). Afterwards, slides were washed in PBS and covered with 4’,6-diamidino-2-phenylindole (DAPI) containing mounting medium (Medipan, Berlin/Dahlewitz, Germany).
For automated γH2AX immunofluorescence microscopy, slides were inserted into the AKLIDES cell damage system and image acquisition as well as analysis were performed as described in detail previously (12,14). In brief, DAPI-stained nuclei were selected in blue fluorescence channel according to morphological criteria using an objective with 60× magnification. After switch into the green fluorescence channel, images of γH2AX foci were obtained of five different focal planes throughout the nucleus. After investigating a minimum of 300 cells per sample, the mean number of γH2AX foci per cell as well as mean γH2AX fluorescence intensity (MFI) of selected nuclei were determined by the analysis software. Cells showing a pan-nuclear staining were recorded separately and samples containing more than 5% pan-nuclear stained PBMCs were excluded from γH2AX foci quantification.
γH2AX analysis using flow cytometry
Preparation of samples for flow cytometric γH2AX detection was performed similarly to slide preparation with minor adaptations according to the protocol by Redon et al. (11). One million etoposide-exposed PBMCs were transferred into a round bottom centrifuge tube and washed in PBS containing 0.5% BSA. After fixation in 1% formaldehyde, cells were incubated in 70% ice cold ethanol over night at 4 °C. Subsequently, samples were washed in PBS containing 0.5% BSA, permeabilized in 0.1% Triton X-100 and blocked in PBS containing 1% BSA. Cell staining was performed at room temperature by applying the γH2AX primary antibody (dilution 1:2,000; 1 h) and additionally the anti-mouse-Alexa-Fluor-488 secondary antibody (dilution 1:500; 1 h).
For flow cytometric analysis, PBMCs were selected according to their forward and side scatter signals using a BD LSRFortessa cell analyzer (BD Biosciences, Mountain View, CA, USA). After measuring a minimum of 20,000 PBMCs, their γH2AX level was quantified by the median fluorescence intensity (MeFI) in arbitrary units (AU) by FlowJo analyzing software (Treestar Inc., Ashland, OR, USA).
γH2AX quantification by immunoblotting
Quantification of H2AX levels by immunoblotting was based on the protocol published by Redon et al. with slight modifications (11). In brief, after etoposide exposure, 1×106 PBMCs were transferred into 1.5 mL tubes, centrifuged at 4 °C and 2,000 g for 5 min and washed in ice cold PBS containing 10 mM NaF. Afterwards, cell pellets were resuspended in 60 µL hot 1× reducing sodium dodecyl sulfate polyacrylamide (SDS) sample buffer and boiled for 10 min at 95 °C. Subsequently, samples were chilled on ice. For SDS gel electrophoresis, lysates were loaded onto a 12.5% SDS gel, following immunoblotting onto a nitrocellulose membrane. After blocking in 5% nonfat dry milk dissolved in 1× Tris buffered saline solution (TBS) for 1 h, the membrane was incubated overnight with anti-phosphohistone H2AX mouse monoclonal IgG primary antibody (Millipore) diluted 1:2,000 in TBS containing 3% BSA. After washing, membrane was incubated for 1 h with 1:10,000 diluted donkey-anti-mouse HRP-conjugated secondary antibody (Dianova, Hamburg, Germany) and washed again. Enhanced chemiluminescence (ECL) substrate (Thermo-Scientific, Rockford, USA) was applied for protein detection. Afterwards, staining was performed analog for β-actin determination. Subsequently, intensity of individual bands was determined with Kodak Image Station quantification software and quantified γH2AX levels were normalized to the corresponding intensity of the β-actin loading control.
Statistics
Data analysis was performed by GraphPad Prism software version 5.01 (Graph Pad Software, La Jolla, CA). Diagrams display the mean and standard error of the mean (SEM) of five independent experiments. Limit of detection (LoD) was determined based on the values obtain from untreated cells, as sum of the mean and three standard deviations. Afterwards, the corresponding etoposide concentration was calculated by linear interpolation.
Results
Using automated fluorescence microscopy, the mean number of γH2AX foci per cell and mean γH2AX intensity per nucleus were obtained. Representative microscopy images are shown in Figure 1A. As also depicted in Figure 1B, increasing etoposide levels induced a rise in γH2AX foci formation detectable even at low concentrations. However, PBMCs exposed to concentrations ≥50 µM exhibited enhanced foci overlap and more than 5% of cells showed pan-nuclear γH2AX staining. Therefore, these data points had to be excluded from γH2AX foci quantification and are marked with X in the diagram.
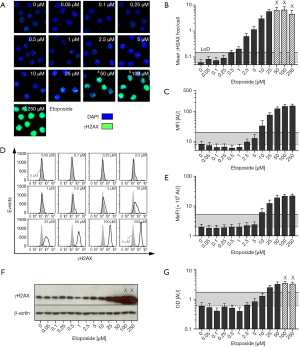
The corresponding dose-response relationship between etoposide concentration and γH2AX MFI obtained by automated fluorescence microscopy is shown in Figure 1C. In contrast to microscopic foci quantification of same images γH2AX intensity values exceeded baseline level only at higher etoposide doses. After a linear increase of γH2AX signal at etoposide concentrations between 2.5–50 µM (R2=0.9862) saturation and enhanced overexposure of γH2AX intensity were observed.
Besides microscopic immunofluorescence analysis, γH2AX intensity of etoposide-exposed PBMCs was also quantified by flow cytometry. Representative histograms and the determined dose-response curve are shown in Figure 1D,E. Similar to the intensity analyses by automated microscopy, flow cytometric γH2AX analysis revealed γH2AX levels elevated above background only at much higher etoposide doses compared to foci quantification. At very high drug concentrations, saturation of the γH2AX signal was observed.
As a third immunological method, we performed immunoblotting to quantify etoposide-induced γH2AX levels, as shown in Figure 1F,G. Similar to immunoassays based on γH2AX fluorescence intensity assessment, a low etoposide concentration did not induce a detectable rise in γH2AX levels. A linear increase of γH2AX expression was observed between a dose range of 2.5–50 µM etoposide (R2=0.9529). PBMCs exposed to etoposide concentrations ≥100 µM had to be excluded from quantification, since the optical density (OD) of the bands exceeded the linear quantification range (indicated with X). But the use of shorter exposure times to avoid overexposure was not sufficient to visualize the weak signals from γH2AX bands of untreated samples.
Additionally, LoD were calculated for each assay as the sum of the mean and three standard deviations obtained from the negative controls. Within diagrams shown in Figure 1 corresponding LoD is indicated as solid line. Performing interpolation the corresponding etoposide concentration was determined. Whereas γH2AX foci quantification revealed a detectable increase in γH2AX signal at etoposide concentrations ≥0.53 µM compared to untreated samples, much higher etoposide doses were necessary to obtain a positive signal for intensity-based measurements (MFI—microscopy: 6.7 µM; MeFI—flow cytometry: 8.7 µM). Also due to high standard deviations, the poorest detection limit was determined for immunoblotting (15.5 µM etoposide).
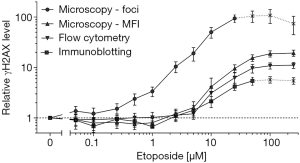
For better comparison of γH2AX levels, values obtained by each method were normalized according to untreated controls. These relative γH2AX data of all four dose-response curves are depicted in Figure 2. In accordance to the LoD, increasing etoposide concentrations induced the strongest gain of the γH2AX signal when assessed by microscopic γH2AX foci quantification.
Discussion
Assessment of γH2AX has been established as one of the most sensitive methods for DNA DSB detection (2). It offers a variety of potential applications in clinics e.g., for monitoring of anti-cancer therapy, determination of radio- or chemo-sensitivity/resistance, for biodosimetry or as biomarker for multiple age-related diseases (2,5-7). But due to several reasons, like the lack of standardization and insufficient data from various multi-center studies, γH2AX analysis has not yet been used as clinical biomarker in laboratory routine.
Among different assays for γH2AX detection, fluorescent microscopic γH2AX foci determination is reported the most sensitive approach (2,9,11). To evaluate the sensitivity of different immunological methods for γH2AX assessment performed in our laboratory, we compared the LoD of automated fluorescent microscopy, flow cytometry and immunoblotting. Therefore, PBMCs were exposed to increasing concentrations of the DSB-inducing cytostatic drug etoposide for one hour.
For automated γH2AX foci quantification, a LoD of 0.53 µM (0.31 µg/mL) etoposide during an exposure of one hour was calculated. In contrast, a more than 10-fold higher etoposide dose was necessary to obtain a detectable increase in γH2AX levels when determined by intensity measurements using either fluorescent microscopy or flow cytometry. Immunoblotting showed the poorest LoD. Further, the comparison of relative increase of γH2AX values after etoposide exposure confirmed the highest sensitivity for γH2AX foci quantification.
In clinical use, plasma etoposide levels of 2–3 µg/mL could be associated with hematological toxicity and antitumor activity was reported at 1–2 µg/mL (15,16). Peak levels may reach up to 18.5 µg/mL and trough levels were reported around 0.2 µg/mL (17). Thus, these levels are either well above the LoD of etoposide determined in this study with regard to antitumor activity or at least equal to the trough levels. In this context, the automated γH2AX foci quantification appears to be a well suited tool for the assessment and follow-up of DNA damage by etoposide in patients as demonstrated for partial body radiation exposure either (18). In contrast, fluorescence intensity analysis of γH2AX formation by flow cytometry or immunoblotting is not applicable for the assessment of antitumor activity due to the at least 10 times higher LoD.
But foci quantification also has limitations. Although single foci can be detected, an underestimation may occur when nuclei show elevated γH2AX levels where individual foci cannot be accurately separated anymore (19). This is also reflected by the flattening of the dose-response curve at etoposide concentrations ≥50 µM. Additionally, an increasing number of cells showed a pan-nuclear staining. Therefore, these samples were excluded form quantification. In cases of high γH2AX expression, evaluation of mean intensity or foci area seems to be more appropriate. In general, protocols of all discussed methods should be adjusted according to respective sample conditions to allow measurement within linear dynamic range.
As shown, γH2AX foci quantification is the best approach when cells with low γH2AX levels are investigated. Besides its high sensitivity, γH2AX fluorescent microscopy furthermore allows assessment of additional parameters. This pertains to cell or foci intensity, size and shape, when combined with image acquisition and image processing. Although foci morphology is often not considered, Watters et al. observed different γH2AX patterns in mouse embryonic fibroblasts depending on the type of genotoxic treatment (20). Further, co-localization studies can be performed applying additional staining techniques, like different DNA damage proteins or chromatin markers (21-25). Besides analysis of single cells, fluorescent microscopy also enables histological tissue examinations of e.g., biopsies or hair follicles (26-29). Especially for research purposes, time-lapse microscopy of γH2AX formation and other repair foci can be performed in living cells by use of fluorescent-tagged proteins or fluorescent antibody fragments (nanobodies) (30-33).
In comparison to fluorescent microscopy, flow cytometry is more suitable for cells expressing more than one γH2AX focus (11) and depending on the intensity level signal strength and dynamic range can be further adjusted by settings of photomultiplier. As a high-throughput technology, flow cytometry enables measurement of several hundred to thousand cells per second and can be combined with additional labelling for DNA content or surface markers. Thereby, it facilitates cell cycle analysis as well as assessment of multiple and even small cell subpopulations. But in contrast to fluorescent microscopy, background intensity derived from autofluorescence or staining variations affect results more strongly and cannot be separated from weak γH2AX foci signals (2). To determine background levels, appropriate staining controls need to be included. Further, new approaches combining analytical characteristics of microscopy and flow cytometry, such as microscope-based laser scanning cytometry or imaging flow cytometry have also been used for γH2AX evaluation (34-37).
In contrast to single cell analysis, immunoblotting- or ELISA-based methods are less sensitive and results are not only dependent on the γH2AX level per cell but are also affected by total cell concentration. Further, apoptotic cells showing pan-nuclear γH2AX expression as well as different subpopulations cannot be distinguished. The ELISA can be performed as classical sandwich ELISA with immobilized capturing antibodies against whole H2A or H2AX molecules and γH2AX-specific detection antibodies. Additionally, modifications have been described utilizing direct plate coating with either cell lysate or whole cells, following fixation and permeabilization, and subsequent detection of γH2AX (38-40). A novel sandwich ELISA developed by Ji et al. does not only determine the level of γH2AX but also of total H2AX to allow quantification of relative γH2AX levels (41).
Immunoblotting experiments of our study were performed with chemiluminescence-based protein determination using ELC-substrate for detection of horseradish peroxidase. Cells treated with high concentrations of etoposide showed an overexposed γH2AX expression, which was above the quantification limit. But when the exposure time was reduced, bands of untreated samples could not be detected anymore. To improve this method, it needs to be investigated whether the substitution of enzyme-coupled secondary antibodies with fluorescence-labeled antibodies can increase the linear quantification range (42). For adjustment of cell concentration, lysates must be aligned according to the cell number or protein concentration. To confirm equal protein loading and to allow normalization of γH2AX levels across the gel, detection of constitutively expressed housekeeping gens, such as β-actin or lamin B1, and even better, total level of H2AX should be included (2,10). In contrast to assays requiring prompt staining of treated cells, lysate-based γH2AX detection by immunoblotting and ELISA has the advantage that a variety of samples can be prepared easily and stored over a certain period of time to allow subsequent and repeated analyses of γH2AX and additional protein levels.
In conclusion, we confirmed that microscopic fluorescent γH2AX foci quantification represents the most sensitive immunoassay for γH2AX determination and is most suitable when γH2AX expression levels are low. Especially, the advancement in automated microscopy may facilitate translation of γH2AX foci analysis into clinical routine since the unsurpassed detection limit allows the assessment and follow-up of antitumor activity of chemotherapeutic substances such as etoposide. Further, improved assay protocols, such as novel ELISA tests as described by Ji et al. (41) should be considered due to their applicability for clinical diagnostics. In general, advantages and disadvantages of each technique should be taken into account to choose the most appropriate assay for the specific problem being investigated.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “DNA Damage Assessment for Precision Medicine”. The article has undergone external peer review.
Conflicts of Interest: The series “DNA Damage Assessment for Precision Medicine” was commissioned by the editorial office without any funding or sponsorship. Dirk Roggenbuck served as an unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Laboratory and Precision Medicine from August 2017 to July 2019. D Roggenbuck is a shareholder of GA Generic Assays GmbH and MEDIPAN GmbH being diagnostic manufacturers. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the local ethics committee and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Blood donors gave written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Behl C, Ziegler C. Cell Aging: Molecular Mechanisms and Implications for Disease. Berlin, Heidelberg: Springer Berlin Heidelberg, 2014.
- Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer 2008;8:957-67. [Crossref] [PubMed]
- Rogakou EP, Boon C, Redon C, et al. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999;146:905-916. [Crossref] [PubMed]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858-68. [Crossref] [PubMed]
- Dickey JS, Redon CE, Nakamura AJ, et al. H2AX: Functional roles and potential applications. Chromosoma 2009;118:683-92. [Crossref] [PubMed]
- Ivashkevich A, Redon CE, Nakamura AJ, et al. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett 2012;327:123-33. [Crossref] [PubMed]
- Redon CE, Weyemi U, Parekh PR, et al. gamma-H2AX and other histone post-translational modifications in the clinic. Biochim Biophys Acta 2012;1819:743-56. [Crossref] [PubMed]
- Reddig A, Rübe CE, Rödiger S, et al. DNA damage assessment and potential applications in laboratory diagnostics and precision medicine. J Lab Precis Med 2018;3:31. [Crossref]
- Rothkamm K, Horn S. gamma-H2AX as protein biomarker for radiation exposure. Ann Ist Super Sanita 2009;45:265-71. [PubMed]
- Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids 2010;2010:1-9. [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Sordet O, et al. γ-H2AX Detection in Peripheral Blood Lymphocytes, Splenocytes, Bone Marrow, Xenografts, and Skin. Methods Mol Biol 2011;682:249-70. [Crossref] [PubMed]
- Runge R, Hiemann R, Wendisch M, et al. Fully automated interpretation of ionizing radiation-induced γH2AX foci by the novel pattern recognition system AKLIDES®. Int J Radiat Biol 2012;88:439-47. [Crossref] [PubMed]
- Willitzki A, Hiemann R, Peters V, et al. New platform technology for comprehensive serological diagnostics of autoimmune diseases. Clin Dev Immunol 2012;2012:284740 [Crossref] [PubMed]
- Willitzki A, Lorenz S, Hiemann R, et al. Fully automated analysis of chemically induced gammaH2AX foci in human peripheral blood mononuclear cells by indirect immunofluorescence. Cytometry A 2013;83:1017-26. [Crossref] [PubMed]
- Miller AA, Tolley EA, Niell HB, et al. Pharmacodynamics of prolonged oral etoposide in patients with advanced non-small-cell lung cancer. J Clin Oncol 1993;11:1179-88. [Crossref] [PubMed]
- Slevin ML, Clark PI, Joel SP, et al. A randomized trial to evaluate the effect of schedule on the activity of etoposide in small-cell lung cancer. J Clin Oncol 1989;7:1333-40. [Crossref] [PubMed]
- Kato Y, Nishimura S, Sakura N, et al. Pharmacokinetics of etoposide with intravenous drug administration in children and adolescents. Pediatr Int 2003;45:74-9. [Crossref] [PubMed]
- Horn S, Barnard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One 2011;6:e25113 [Crossref] [PubMed]
- Schmid TE, Zlobinskaya O, Multhoff G. Differences in Phosphorylated Histone H2AX Foci Formation and Removal of Cells Exposed to Low and High Linear Energy Transfer Radiation. Curr Genomics 2012;13:418-25. [Crossref] [PubMed]
- Watters GP, Smart DJ, Harvey JS, et al. H2AX phosphorylation as a genotoxicity endpoint. Mutat Res 2009;679:50-8. [Crossref] [PubMed]
- Balajee AS, Geard CR. Replication protein a and γ-H2AX foci assembly is triggered by cellular response to DNA double-strand breaks. Exp Cell Res 2004;300:320-34. [Crossref] [PubMed]
- Marková E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol 2007;83:319-29. [Crossref] [PubMed]
- Cowell IG, Sunter NJ, Singh PB, et al. Gamma-H2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One 2007;2:e1057 [Crossref] [PubMed]
- Kim JA, Kruhlak M, Dotiwala F, et al. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J Cell Biol 2007;178:209-18. [Crossref] [PubMed]
- Rübe CE, Lorat Y, Schuler N, et al. DNA repair in the context of chromatin: new molecular insights by the nanoscale detection of DNA repair complexes using transmission electron microscopy. DNA Repair (Amst) 2011;10:427-37. [Crossref] [PubMed]
- Rübe CE, Grudzenski S, Kühne M, et al. DNA double-strand break repair of blood lymphocytes and normal tissues analysed in a preclinical mouse model: Implications for radiosensitivity testing. Clin Cancer Res 2008;14:6546-55. [Crossref] [PubMed]
- Schuler N, Rübe CE. Accumulation of DNA Damage-Induced Chromatin Alterations in Tissue-Specific Stem Cells: The Driving Force of Aging? PLoS One 2013;8:e63932 [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Zhang YW, et al. Histone γH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res 2010;16:4532-42. [Crossref] [PubMed]
- Redon CE, Dickey JS, Nakamura AJ, et al. Tumors induce complex DNA damage in distant proliferative tissues in vivo. Proc Natl Acad Sci U S A 2010;107:17992-7. [Crossref] [PubMed]
- Jakob B, Splinter J, Durante M, et al. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci U S A 2009;106:3172-7. [Crossref] [PubMed]
- Li W, Li F, Huang Q, et al. Quantitative, noninvasive imaging of radiation-induced DNA double-strand breaks in vivo. Cancer Res 2011;71:4130-7. [Crossref] [PubMed]
- Rajan M, Mortusewicz O, Rothbauer U, et al. Generation of an alpaca-derived nanobody recognizing γ-H2AX. FEBS Open Bio 2015;5:779-88. [Crossref] [PubMed]
- Georgescu W, Osseiran A, Rojec M, et al. Characterizing the DNA Damage Response by Cell Tracking Algorithms and Cell Features Classification Using High-Content Time-Lapse Analysis. PLoS One 2015;10:e0129438 [Crossref] [PubMed]
- Zhao H, Albino AP, Jorgensen E, et al. DNA damage response induced by tobacco smoke in normal human bronchial epithelial and A549 pulmonary adenocarcinoma cells assessed by laser scanning cytometry. Cytometry A 2009;75:840-7. [Crossref] [PubMed]
- Tanaka T, Halicka D, Traganos F, et al. Cytometric analysis of DNA damage: phosphorylation of histone H2AX as a marker of DNA double-strand breaks (DSBs). Methods Mol Biol 2009;523:161-8. [Crossref] [PubMed]
- Furia L, Pelicci PG, Faretta M. A computational platform for robotized fluorescence microscopy (II): DNA damage, replication, checkpoint activation, and cell cycle progression by high-content high-resolution multiparameter image-cytometry. Cytometry A 2013;83:344-55. [Crossref] [PubMed]
- Bourton EC, Plowman PN, Zahir SA, et al. Multispectral imaging flow cytometry reveals distinct frequencies of γ-H2AX foci induction in DNA double strand break repair defective human cell lines. Cytometry A 2012;81:130-7. [Crossref] [PubMed]
- Audebert M, Riu A, Jacques C, et al. Use of the γH2AX assay for assessing the genotoxicity of polycyclic aromatic hydrocarbons in human cell lines. Toxicol Lett 2010;199:182-92. [Crossref] [PubMed]
- Matsuzaki K, Harada A, Takeiri A, et al. Whole cell-ELISA to measure the γH2AX response of six aneugens and eight DNA-damaging chemicals. Mutat Res 2010;700:71-9. [Crossref] [PubMed]
- Johnston ML, Young EF, Shepard KL. Whole-blood immunoassay for γH2AX as a radiation biodosimetry assay with minimal sample preparation. Radiat Environ Biophys 2015;54:365-72. [Crossref] [PubMed]
- Ji J, Zhang Y, Redon CE, et al. Phosphorylated fraction of H2AX as a measurement for DNA damage in cancer cells and potential applications of a novel assay. PLoS One 2017;12:e0171582 [Crossref] [PubMed]
- Mathews ST, Plaisance EP, Kim T. Imaging systems for westerns: chemiluminescence vs. infrared detection. Methods Mol Biol 2009;536:499-513. [Crossref] [PubMed]
Cite this article as: Reddig A, Roggenbuck D, Reinhold D. Comparison of different immunoassays for γH2AX quantification. J Lab Precis Med 2018;3:80.


