
Molecular biomarkers of DNA damage in diffuse large-cell lymphoma—a review
Introduction
Cancer is the second most frequent cause of death worldwide according to the World Health Organisation (WHO) (1). One of the most aggressive forms of Non-Hodgkin’s lymphoma, diffuse large B-cell lymphoma (DLBCL), is diagnosed in more than 50% of lymphoma patients over the age of 65 (2). As for most malignant diseases, the risk of developing DLBCL increases with age, and more than 40% of patients eventually succumb to their disease (3,4). In recent years, therapy for many cancers and leukaemia’s has been fundamentally improved based on a deeper understanding of the molecular hallmarks underlying the disease, paving the way for the concept of personalized medicine both with regard to therapy selection and response monitoring. As cancer is basically a genetic disorder, most malignant diseases arise from one sort or another of de novo deoxyribonucleic acid (DNA) damage, be this point mutations or chromosomal rearrangements. At the root of these lies unrepaired DNA damage, which occurs frequently under physiological conditions as the DNA is exposed to damaging agents, such as reactive oxygen species (5) or ultra-violet (UV) irradiation (6).
The most severe form of DNA damage is a DNA double-strand break (DSB). Caused by chemical or physical noxae, it leads to genomic instability (7,8) and thus increases the probability of cancerogenesis (9). Apart from cancer, DNA damage is related to diseases associated with aging such as Xeroderma pigmentosum, Cockayne’s syndrome or Trichothiodystrophy (10). Some mutations in oncogenes or tumour suppressor genes, such as of the protein 53 (p53) significantly increase the incidence of cancer in patients. For example, mutations of the p53 gene leading to inactivation of the protein can be detected in 50% of all human tumours such as for instance breast, lung, liver tumours or lymphomas, but only in a small percentage of the healthy population (11).
Currently, cancer is often diagnosed by radiological examinations or biopsies, and for a complete diagnosis consisting of grading and staging, both are usually required. As highly proliferative malignant cells run in hypermetabolic state, they can also be detected by radioactively labelled metabolic substrates in conventional scintigraphy or positron emission tomography (12,13). All these procedures are often stressful for the patient, carry the risk of injuries and adverse effects, and have a varying degree of sensitivity and specificity, which lead to diagnostic insecurity and may necessitate repeated procedures, for instance in the case of a negative tissue biopsy that is clinically highly suspicious for cancer.
A diagnostic “magic bullet” would therefore be highly desirable, as is sought for with the emerging concept of liquid biopsies, intended to replace tissue biopsies with materials from blood samples, such as soluble DNA, peripheral blood mononuclear cells (PBMC), and circulating tumour cells, for diagnosis and monitoring of malignant disease (14). Circulating tumour cells predict the probability of a poor prognosis in early stages of breast cancer and may thus give a valuable contribution to decide whether or not upon initial surgery a woman should receive adjuvant therapy. Rack et al. showed that patients with at least five circulating tumour cells per 30 mL of blood have the worst prognosis and survival probability [hazard ratio (HR) for disease-free survival =4.51, 95% confidence interval (CI): 2.59 to 7.86; HR for overall survival (OS) =3.60, 95% CI: 1.56 to 8.45] and that the detection of circulating tumour cells after chemotherapy correlates with the risk of relapse (15). However, the clinical meaning of these findings is subject to clinical trials (16,17), and currently it is unclear where and how this prospective information can be used to actually improve prognosis. Technically, the liquid biopsy approach relies on biomarkers that are clinically meaningful (18) and can be easily assessed with good sensitivity and specificity (19), preferably in multiparametric screening systems.
In this review we discuss potentially relevant biomarkers for diagnosis, prognosis and prediction of therapy response in malignant diseases. We will first discuss the current genetic understanding of DLBCL and the repair systems that normally prevent a given genetic damage to manifest as malignant disease. We will then focus on DNA DSBs and biomarkers of it such as the p53 binding protein (53BP1), the histone variant 2AX (H2AX), and the kinases Ataxia telangiectasia mutated protein (ATM), Ataxia telangiectasia and Rad3-related protein (ATR), and DNA-dependent protein kinase (DNA-PK). This will be followed by eight other potential biomarkers associated with either cancer prevention or induction: Myc, B-cell lymphoma 2 (BCL-2), Breast cancer 1 (BRCA1), signal transducer and activator of transcription 3 (STAT3), 8-Hydroxy-2'-desoxyguanosine (8-OHdG), recombinase Rad51, proteasome activator 28γ (PA28γ), and tumour suppressor p53, which have all been suggested for monitoring and predicting the outcome of therapies in cancer patients, in particular those with DLBCL.
Diffuse large B-cell lymphoma
DLBCLs are the most common type of aggressive lymphomas with a crude incidence of 3.8 per 100,000 in Europe diagnosed in 2000 to 2002 (20). In fact, they comprise a heterogeneous entity with the common feature of large B-cells arranged in diffuse patterns. Morphologically an immunoblastic, centroblastic, and anaplastic type can be distinguished (21,22) and genetically they can be roughly categorized into the germinal centre B-cell (GCB) and the activated B-cell (ABC) subtype (22) by their genetic signatures. Their characterization by flow cytometry or immunohistochemistry relies on different pan-B-cell antigens such as CD19, CD20 and CD22 as well as on B-cell transcription factors Paired box protein 5 (Pax5) and Organic cation transporter 2 (OCT2) (21). Untreated it is almost always lethal within a few weeks to months, but the introduction of combination chemotherapy with cyclophosphamide, doxorubicin, vincristine and prednisone (CHOP) some 40 years ago, has by itself brought in long-term remissions and a considerable cure rate. Since then, the addition of the anti-CD20 monoclonal antibody rituximab by the end of the 1990’s has further improved outcomes to response rates of 40% to over 90% and 5-year survival rates of 35% to 85%, depending on age and clinical risk group (3,23,24). The R-CHOP protocol has since been the gold standard of therapy which more aggressive therapy regimens failed to improve upon. Treatment options for patients not responding to R-CHOP or experiencing a relapse are scarce and prognosis for these patients is poor.
Patients harbouring a GCB lymphoma are more likely to survive at least 5 years upon R-CHOP therapy than those with the ABC subtype (69%±3% for GCB vs. 53%±5% for ABC vs. 60%±4% for unclassified cases; P=0.02 for GCB vs. ABC) (25). Nonetheless, 30% to 40% of all DLBCL patients relapse after R-CHOP therapy (3,4). Response assessment relies upon restaging by computed tomography (CT) alone or its combination with fluorodeoxyglucose positron emission tomography (FDG-PET; for the combination, PET-CT). Both are highly sensitive, and PET-CT is much more specific in distinguishing residual vital tumour tissue from fibrotic scars (26). Earlier detection of a relapse does not seem to translate into better survival and radiological follow-up frequency has recently been drastically reduced.
Hence, to improve the prognosis for patients not cured with standard R-CHOP, a better molecular understanding of DLBCL development and progression is a predominant focus of research, and significant biomarkers accessible for liquid biopsies appear to promise great gains for DLBCL patients (27).
Cellular repair mechanisms of DNA damage
For apparent reasons, in multicellular organisms cell cycle and DNA-replication are tightly controlled and a multitude of highly conserved, evolutionary ancient control and repair mechanisms usually identifies and corrects errors or submits the cell to apoptosis (28). As a result, if an error is detected, the cell cycle halts and repair mechanisms attempt to restore the original DNA sequence. Once this is successful, the cell proceeds into the next phase of the cell cycle (29). If the DNA cannot be repaired, the cell either enters senescence (30), necroptosis (31), or apoptosis, all of which maintain genetic integrity. Only if this fails, too, the irreparable DNA damage is passed on to the next generation (32). The latter event can lead to loss of protein function or, over the course of further generations, to the accumulation of additional mutations and ultimately to the uncontrolled cell proliferation that defines cancer (29,33).
The main repair systems in mammalian cells are base excision repair, nucleotide excision repair, mismatch repair and DSB repair (28). Damage at a single nucleotide base (e.g., loss or deamination), including chemically oxidized or methylated bases such as 3-methylguanine or 8-OHdG, can be repaired by base excision repair, a mechanism present in most organisms (34). In patients with malignant tumours, high levels of 8-OHdG were found (35-37), marking the critical importance of the base excision repair system.
DNA damage involving complete nucleotides such as UV-induced pyrimidine dimers or bases modified by polycyclic aromatic compounds (10) is corrected by nucleotide excision repair. The nucleotide excision repair system is closely linked to transcription-coupled repair and DNA interstrand crosslink repair (38). A congenital defect of the nucleotide excision repair system is associated with Xeroderma pigmentosum and Trichothiodystrophy (39). Xeroderma pigmentosum patients are hypersensitive to UV irradiation and at increased risk for malignant skin neoplasms (40).
Replication errors, such as mismatched or missing bases, are corrected by mismatch repair. Major DNA damage and DSB can be corrected by homologous recombination (HRe) or non-homologous end joining (NHEJ). Defects in genes involved in HRe are found in about 50% of all epithelial ovarian cancers (41), and 15% of those affected carry germ line mutations of BRCA1 and BRCA2 (42), the genes for two proteins that interact with Rad51 in repairing DSB via HRe (43) and thus demonstrate these mechanisms’ importance for preventing cancer. It is thought that during mitosis, HRe is regularly employed in the late S and G2 phases when the sister chromatids can serve as templates, whereas NHEJ is more prominently active in G1 and early S phase (44) and repairs the majority of DNA DSBs (45-47).
DNA double strand breaks and cancer
Development and repair of DNA DSB
DNA DSBs are considered the most harmful form of DNA damage as they can promote genomic instability and increase the risk of cancer (7,33,48-51). The repair of DNA DSBs is more complex than that of single nucleotide damages because the correct anti-parallel strand is no longer available as a template (35,38,49,52). Therefore DSBs cannot be repaired through a simple ligation (49), but HRe and NHEJ need to compensate for the loss of both DNA strands (7,48,49,53). The complex machinery of histones and enzymes involved in this process also offers the methods that have been proposed to detect and quantify DNA DSBs (54). Histone modifications associated with DNA damage have attracted growing interest for early cancer detection in recent years.
The histone 2AX (H2AX) has become a widely used biomarker for DNA DSBs (51,55-57). Upon the occurrence of a DSB, H2AX is phosphorylated at serin 139 (then termed γH2AX) by members of the phosphoinoside-3 kinase (PI-3K) family and binds to the damaged DNA (Figure 1) (55,56). The formation of γH2AX is easily detected using antibodies against the phosphorylated variant, which can be localized as discrete nuclear foci (57-59).
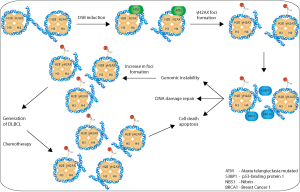
The phosphorylated histone 2AX variant is associated with interleukin 1α, which is involved in inflammation, microbial diseases and cancer and influences tumours by promoting proliferation, the expression of chemokines, adhesion molecules, and pro-survival factors. During stress such as UV irradiation, interleukin 1α is secreted by necrosis or pyroptosis (60) and co-localizes with γH2AX and histone deacetylase-1 at sites of DNA damage. The function of this nuclear localization is not known so far, but localization with biomarkers such as 53BP1 (61-64), BRCA1 (65,66) and Rad51 (66) was also observed for γH2AX. Those co-localizations indicated an association of γH2AX with DNA damage repair, cell cycle progression and HRe.
Increased γH2AX foci formation has been observed in a wide variety of tumours and meanwhile has been investigated in more than 35 clinical studies [reviews in (67,68)]. Its prognostic relevance has been shown for different tumours such as triple negative breast cancer (P=0.01; HR =4.06, 95% CI: 1.40–11.77) (69). Incubation of PBMCs with different chemotherapeutic drugs such as etoposide, rapamycin (57) or doxorubicin (70) as well as irradiation (51,71-74) induces γH2AX formation (51,57,70,75). Hence it has been suggested that cytotoxic chemotherapy may further increase γH2AX foci formation in cancer patients, who already experience more DNA DSBs than healthy donors (Figure 1). Interestingly, γH2AX foci also appear to increase consistently with exercise (P=0.04 to <0.001) as observed with runs ranging from 5 to 42 km (P<0.001) (76).
In addition to the promising application possibilities, there are also controversial concerns about the use of H2AX as a biomarker. These are based primarily on the high heterogeneity of tumour diseases. It is also debatable whether ex vivo analyses really have the same significance as in vivo analyses. There is also evidence that results of γH2AX foci assay depends on the experimental settings (77). High heterogeneity would also be expected in in vivo samples due to uncontrollable factors such as nutritional intake, oxygen supply or tumour microenvironment. However, a study by Rassamegevanon et al. demonstrated a high grade of heterogenetic results when human head and neck squamous cell carcinoma models were treated with ionising radiation ex vivo, compared to in vivo. This mean that the transfer of a tumour into a new microenvironment could increase the degree of heterogeneity in H2AX foci (77). Whether this is also the case for liquid biopsies such as the isolation of PBMCs is subject to ongoing debate.
Regarding DLBCL in particular, Derenzini et al. found phosphorylated H2AX in 47% of patients where it appeared to predict 5-year OS, which was 41% in γH2AX positive as opposed to 70% in γH2AX negative patients (P<0.01) (68). While these data need to be verified in larger cohorts, they suggest a potential prognostic value of this parameter and might in addition point at a novel therapeutic target as the authors suggest.
Involvement of DNA kinases
Phosphorylation of H2AX can be catalysed by three kinases: the serine protein kinase ATM, the phosphatidylinositol 3-kinase-related kinase DNA-PK, and the serine/threonine protein kinase ATR (78,79). Defects in ATM increase cancer incidence in patients and can lead to the eponymous Ataxia telangiectasia syndrome which is associated with increased radiation sensitivity (80-82). Upon DSB induction, ATM phosphorylates downstream targets such as p53, mouse double minute 2 homolog (Mdm2), checkpoint kinase 1 and 2 (Chk1 and Chk2), BRCA1 and nibrin next to H2AX and thus has a variety of influences for instance on apoptosis, cell cycle checkpoints and DNA repair (33,48,83). The exact function of ATM-mediated phosphorylation remains unclear (79,84). However, γH2AX foci formation is strongly reduced in cells which lack DNA-PK and reduced ATM levels (66).
In DLBCL patients, ATM mutations have been found to be associated with inactivation of the tumour suppressor gene ARF and tumor protein P53 (TP53). Missense variants of ATM have been discovered and are of unclear functionality. Nonetheless, it was suggested that mutations in ATM and inactivation of the ARF-TP53 pathway may promote DLBCL (85). As loss of its function appears to lead to genomic instability, cancer susceptibility and profound sensitivity to ionising radiation (86,87), ATM is a potential target for cancer therapy. On that account, several ATM inhibitors are of interest such as Wortmannin (88), KU-55933 (89) or the most potent ATM-inhibitor KU-559403 (90), although none of these is currently in clinical development (Table 1) (87). Among these, KU-559403 stands out for its improved bioavailability and solubility (90). In a small clinical study, 55% of DLBCL patients had a mutation of ATM in addition to TP53 (P=0.015, n=9), which suggests a non-random connection (85). The ATM kinase can be detected by immunoblotting and has been shown to be an additional diagnostic tool for Ataxia Telangiectasia besides the colony survival assay (100).
Table 1
| Potential Biomarker | Target structure | Detection Method(s) | Clinical relevance | ClinicalTrails.gov Studies | Additional References |
|---|---|---|---|---|---|
| 53BP1 | Protein, DNA | IIF, FISH | Poor diagnostic value in MS (63), correlation with molecular grouping in triple breast cancer (91), poor prognosis in triple negative breast cancer (69), foci formation after irradiation in mesenchymal stem cells (92) | 6 | (63,92,93) |
| 8-OHdG | Protein | IIF, HPLC, ELISA, IHC | Quantification of oxidative stress (94-97), prognostic marker for lymphoma (98) | 32 | (97,99) |
| ATM | Protein | IIF, WB | Mutation contribute to DLBCL (85) | 103 | (100-102) |
| ATR | Protein | IIF | Inhibitor used in clinical studies (87) | 36 | (103) |
| BCL-2 | DNA, Protein | FISH, IHC | Risk-stratify of DLBCL patients (104), predict recurrence of prostate cancer after radical prostatectomy (105), prognostic marker in breast cancer (106), inferior survival of DLBCL (107), phase-II study of inhibitor (108) | 219 | (104) |
| BRCA1 | Protein, DNA | WB, FISH, IIF | Mutations in BRCA1 also indicated high occurrence of γH2AX in breast cancer (69), promoter hypermethylation associated with more aggressive breast cancer profile (91), cisplatin chemotherapy high activity in breast cancer with BRCA1 mutation (109,110), BRCA1 level determines OS in lung carcinomas (111), higher breast cancer risk for females with BRCA1 mutation (112,113) | 334 | (114,115) |
| Myc | DNA, Protein | FISH, IIF, IHC, qPCR | Poor prognosis for DLBCL (107,116), risk-stratify of DLBCL patients (104,117), prognostic value for OS in DLBCL (118), Myc rearrangement strongly adverse prognostic factor (119), pSTAT3 expression associated with Myc (120) | 99 | (104,114,121-123) |
| DNA DSB (γH2AX) | Protein | IIF, FISH, WB, FACS | Poor diagnostic value in MS (63), poor prognosis in triple negative breast cancer (69), γH2AX increase during exercise (76), increased in CTC and PBMCs after treatment with PARP-inhibitor (124), prediction of OS in DLBCL (68), quantitative biomarker for low-dose radiation (72), increased foci number in chronic lymphocytic leukemia (125), foci formation after irradiation in mesenchymal stem cells (92) induction after treatment with aza-cytosine nucleosides (126) and radiotherapy (127) implies integral body dose (128), γH2AX declines with increased age (129), biomarker for DNA damage in TP53 mutated high-risk endometrial cancer (130) | 37 | (55,92,131-133) |
| DNA-PK | Protein | WB, IIF | Cytotoxicity induction and inhibition of chronic lymphocytic leukaemia survival signals through DNA-PK inhibitors (134), high-risk endometrial cancer deficient for DNA damage response biomarkers (130) | 7 | (102,132) |
| p53 | Protein, DNA | IHC, HPLC, FISH, DNA Microarray | Predict recurrence of prostate cancer after radical prostatectomy (105), associated with increased tumour size of breast cancer patients (size expressed as binary variable) (106), higher amount of DNA damage in TP53 mutated high-risk endometrial cancer (130), Predicted output of DLBCL (135), mutation of TP53 correlate with poor OS in DLBCL (117,136) | 362 | (114,117,136,137) |
| PA28γ | Protein | WB, ELISA | Used in one clinical study depending neoplasms (138) | No study found | (139,140) |
| Rad51 | Protein | IIF, WB | Foci formation after prolongs irradiation in mesenchymal stem cells (92), trend for improved survival in high-risk endometrial cancer and positive correlation with γH2AX (130), low Rad51 scores correlated with high histological grade of breast cancer and predict chemotherapy response (141), co-localization with BRCA1 (66) and 53BP1 (130) | 44 | (142-144) |
| STAT3 | Protein | IHC, DNA Microarray | Clinical trial target for DLBCL (120), pYSTAT3 predicted outcome of DLBCL and is associated with poor OS (135,145,146) | 64 | (146) |
Biomarkers are listed with target structure and detection methods. Studies on ClinicalTrail.gov were found by searching for Cancer and the specific biomarker. ELISA, enzyme-linked Immunosorbent Assay; FACS, fluorescence-activated cell scanning; FISH, fluorescent in situ hybridisation; HPLC, high performance liquid chromatography; IHC, immunohistochemistry; IIF, indirect immunofluorescence; qPCR, quantitative Polymerase chain reaction; WB, Western Blot (Immunoblotting).
Defects of ATR, too, increase radiation sensitivity and reduce monitoring of cell cycle checkpoints. This kinase phosphorylates proteins such as H2AX (65), Chk1 (93) and BRCA1 (147) and helps to activate the cellular tumour antigen p53 (148). Therefore, ATM and ATR play a central role in the cellular response to radiation, and their functions overlap. They influence each other through mutual expression regulation (131,132,149) and phosphorylation (150,151). Potent inhibitors of ATR are NU6027 (152), VE-821 (133) or AZD6738 (87). The latter is reportedly being examined in a clinical study (ClinicalTrials.gov: NCT02223923), which is currently suspended, however (Table 1) (153). Biomarkers to monitor the effects of ATR and/or ATM inhibitors have not been established. Suggestions include p53 and activated oncogenes such as Myc, pTP53, γH2AX, as well as ATM or phosphorylated ATM itself (87). Immunostaining of ATR was performed by Jazayeri et al. and showed that ATM and ATR work in the same signalling pathway upon irradiation, with ATM functioning upstream of ATR (Table 1) (103). Moreover, co-localization of ATM and γH2AX foci at sites of DSBs has been shown in Hela cells (101).
DNA-PK is an enzyme complex consisting of the three components Ku80, Ku70, and the catalytic subunit DNA-PKcs (DNA-dependent protein kinase catalytic subunit), and is involved in NHEJ. This complex only assembles immediately on the DNA to be repaired. Ku70/Ku80 initially binds as an annular heterodimer to the broken DNA double strand ends. Subsequently DNA-PKcs joins this heterodimer, leads to a change in conformation and thus activates the catalytic subunit (45,154,155). Two DNA-PK complexes are able to mediate joining of DNA at opposite ends, prevent inadvertent loop formation and reassemble the two strands (156). Knockout mouse embryonic fibroblast cells lacking this complex were more susceptible to damage from ionizing irradiation and showed slower DSB repair with prolonged cell cycle arrest. Lack of DNA-PK seemed to affect overall DNA damage response only little, but to disrupt severely the repair of DSB (102). Formation of γH2AX foci upon irradiation was similar in DNA-PKcs deficient and wild type mouse cells, but could neither be detected in ATM-deficient cells nor after exposure to high doses of the phosphatidylinositol 3-kinase inhibitor Wortmannin. In conclusion, in the absence of ATM, DNA-PK appears to maintain a low level of H2AX phosphorylation (78). Hence it is probably directly involved in H2AX phosphorylation and foci formation at the site of DSBs. DNA-PK foci themselves can be detected by indirect immunofluorescence (IIF) (132) and offer another marker for DNA-DSBs amenable to automated immunofluorescence imaging (Table 1).
53BP1 as diagnostic biomarker for DSB repair
The DNA damage checkpoint protein p53 binding protein 1 (53BP1) accumulates rapidly with γH2AX at the site of DNA DSBs (61,62) and is itself a substrate for ATM-dependant phosphorylation. The protein appears to become physiologically phosphorylated to some low degree, but hyperphosphorylation was detected only upon ionizing irradiation (62). In DLBCL a single copy loss in the 53BP1 locus on chromosome 15q15 was found (157), and this haploinsufficiency may be a predisposing event in some DLBCL. 53BP1, too, plays a crucial role in DSB repair (61,62,157). H2AX is not necessary to recruit 53BP1, but is needed for maintaining and increasing 53BP1 levels at a DSB site (158). Upon a noxa, the protein was shown to co-localize with γH2AX (61-64) and BRCA1 (65). Moreover, quantitative γH2AX levels correlated with 53BP1 (P=0.053) and BRCA1 (P=0.011) in breast cancer cell lines (69). In triple negative breast cancer, 53BP1 protein expression correlated with molecular grouping of unmethylated BRCA1 promoter (53% of unmethylated vs. 27.8% of methylated cancers, P=0.045) (91). As opposed to these malignant diseases, in multiple sclerosis (MS) neither γH2AX nor 53BP1 foci analysis differed from healthy donors (for both biomarkers P=0.6) (63). However, 53BP1 is involved in DNA damage-response pathways (159) and enhances p53-dependent transcription (160). Since 53BP1 is considered a marker for the repair of DNA DSB, its use together with γH2AX, which is a marker for the early detection of DSB (Table 1), would be advisable.
Detection of DNA DSB and their application as diagnostic biomarker
Various alternative methods are employed to detect DNA DSB. The classical method is the single cell gel electrophoresis or comet assay (so named after the migration pattern of DNA) of Östling and Johansson (161). But 100 times greater sensitivity can be achieved by flow cytometry (162). Using γH2AX as a common biomarker for DNA damage has since become popular. Immunoblots are conventionally used to detect the histone in total protein lysates. However, besides its lower sensitivity compared to immunofluorescence (57), immunoblotting is time-consuming and ill-suited for larger sample numbers in a clinical setting (7,163). To counter these drawbacks, fully automatized IIF platforms have been developed and adapted specifically for the analysis of human lymphocytes (also summarized in Table 1) (51,57,70,164-173). Equipped with a computerized image analysis, an automated IIF platform demonstrated superior sensitivity compared to fluorescence microscopy, flow cytometry or immunoblotting on PBMC treated with etoposide (174). Experiments showed a dose-dependent increase of the γH2AX damage response as a consequence to cytotoxic treatment (51,57). The advantage of automated IIF is the detection of individual foci per cell (count data), nucleus size (diameter) and fluorescence intensity (57). As opposed to flow cytometry and manual fluorescence microscopy, automated microscopy allows for the capture of multiple z-planes in micrometre distances and subsequent three-dimensional analysis (64). In addition, several fluorescence colours can be integrated into one multiplexing analysis, limiting the number of cell structures to be evaluated simultaneously only by the available fluorescence dyes. In our group, we have analysed the influence of cryopreservation of human PBMCs with this technique. Surprisingly in the light of cell therapies using the same cryoconservation technology, we detected a significant decrease in γH2AX levels after cryopreservation (175).
Fluorescent in situ hybridization (FISH) is a widely applied and highly sensitive method for detecting specific nucleic acid sequences through the specific binding of labelled DNA or RNA probes (114,176). Combining γH2AX fluorescence microscopy with FISH offers a way to characterize the DNA actually affected by detected DSBs.
Nakamura et al. showed that telomere damage highly contributed to overall endogenous DNA double-strand damages in tumour cells (177). Telomere-specific FISH probes were used to differentiate telomeric from extra-telomeric DNA damage. Combined analysis of γH2AX foci and FISH allowed to discriminate DNA double-strand ends from DSBs at chromosomal ends, e.g., within telomeres. This study found that γH2AX foci were directly related to the extent of telomere damage and inversely related to telomerase activity (177).
Brugat et al. used FISH for the detection of complex chromosomal aberrations in chronic lymphocytic leukaemia cells known for telomere dysfunction (125) and showed that critically short telomeres activate the same response as DSB, characterized by foci of Telomeric repeat-binding factor 2 and γH2AX and/or 53BP1 as well as proteins of the NHEJ repair system. By analysing telomere and other frequent chromosomal deletions in metaphase chronic lymphocytic leukaemia cells, they demonstrated an association of short telomeres with genomic instability (125).
FISH was also successfully applied to investigate the role of H2AX in B-lymphocytes during the recombinatorial switch in immunoglobulin heavy chain class (178,179). Using FISH probes targeting different regions of the heavy chain genes, Franco et al. and Yin et al. proposed a general role of the DSB response in promoting end joining and preventing chromosomal breaks or translocations through chromatin modifications performed by H2AX (178,179).
As mentioned earlier, while DSBs may cause cells to become genetically instable and then cancerous, the vast majority will first halt the cell cycle and then lead to apoptotic cell death (Figure 1). Some cytotoxins utilize the lethal effect of DSBs by either inducing them (etoposide) or by provoking malfunctions of critical enzymes such as topoisomerase II (anthracyclines, irinotecan) (57,70). The DNA-DSB response factors γH2AX, 53BP1, ATR, ATM, and DNA-PK could therefore be used to monitor cancer treatment and possibly predict its outcome from PBMC.
The proto-oncogenes Myc and BCL-2 as diagnostic markers
To get a broader picture in the development of DLBCL, other biomarkers should be considered. The proto-oncogene Myc and the anti-apoptotic protein BCL-2 are both involved in the regulation of apoptosis. The overexpression of Myc lowers the level of BCL-2 and promotes apoptosis (180). In most oncological cases, increased expression of Myc however results in overexpression of BCL-2, which favours anti-apoptosis in cancer cells (181). Therefore, Myc and BCL-2 are key players in inducing oncological diseases.
Lymphomas with rearrangements of the proto-oncogene Myc (also known as c-Myc) carry a poor risk even with CHOP or R-CHOP therapy (182). There are two controversially discussed types of FISH probes for Myc rearrangements, break-apart probes and dual-fusion probes (119,182), of whom the break-apart probes appear more efficient (183). Some DLBCL patients show Myc overexpression without gene alterations: existing data suggest that only one third of patients with Myc overexpression actually carry a translocation (104,121), while other authors maintain that these be false positives and should be re-evaluated by FISH screening (Table 1) (122).
Myc was identified as a biomarker for DLBCL in a meta-analysis comprising 4662 cases. The HR of patients with c-Myc abnormalities compared to DLBCL patients without, calculated and analysed from 19 different studies, was situated at 2.22 (95% CI, 1.89 to 2.61) (184). The majority of studies in this meta-analysis found Myc predictive for the outcome of DLBCL (184).
The oncogenic effect of Myc depends rather on mutations influencing its quantitative expression than on qualitative alterations of its fundamental functions (185). DNA breaks within the myc gen can be detected by quantitative PCR, but no correlation between Myc DNA breaks and high mRNA levels was found. Immunohistochemistry (IHC) Myc staining correlates with myc transcript numbers. For a precise detection of several Myc mutations, multiple or multiplex Polymerase chain reaction (PCR) would be necessary (122). But the optimal method for the detection of myc expression still seems to be controversial.
It is suggested that Myc is associated with nearly half of all human oncological diseases (185). In ovarian cancer, patients expressing high levels of Myc mRNA lived shorter with earlier relapses (186). Along the same line, high myc expression indicated poor overall survival in DLBCL (123).
In addition to these findings, Johnson et al. showed that BCL-2 determines the outcome of Myc-positive DLBCL. The so-called double-hit (Myc-positive and BCL-2-positive) phenotype has been confirmed as a potent predictor of poor risk (104). Detection of BCL-2 and Myc protein expression by IHC is cheaper, faster, and equally reproducible compared to the detection of Myc translocations by FISH (104). Molecular hallmarks such as BCL-2 or the ABC type deteriorate the already poor outcome, as well as R-CHOP treatment in elderly patients (187-190). DLBCL patients are BCL-2 positive in 66% of cases and responded better to R-CHOP therapy than to a therapy without rituximab. Those patients also indicated a better overall survival (67%±9% versus 48%±11%, P=0.004) than patients positive for BLC-2 treated with CHOP alone (189). In addition, the frequency of Myc positive cases increases from 35% to 62% when γH2AX negative cases are compared to positive patients (P=0.02) (68), making it a more likely candidate then BCL-2.
BRCA1 as biomarker for breast cancer and lymphoma
Mutations in the tumour suppressor gene BRCA1 (also termed breast cancer susceptibility gene 1) are predictive for highly increased risks of breast and ovarian cancer, and about 5% of all breast cancer cases can be attributed to BRCA1 mutations. However, its tumour suppressor function is ubiquitous and failure of it is also involved in lymphoma (92). The median survival in a low BRCA1 group of non-small-cell lung cancer patients was 11 month (95% CI: 1.1 to 20.9) and two-year survival rate was 41.2% compared to the high BRCA1 group with same median survival (95% CI: 8.2 to 13.8), but a two-year survival rate of 0% (94). BRCA1 is associated with genotoxic stress and γ-irradiation (95,96). Increased γH2AX levels were detected in BRCA1-mutated breast cancer cell lines (P=0.011) (69). Phosphorylation of H2AX has been shown to be a recruitment signal for BRCA1 (66). Upon DNA damage, γH2AX and BRCA1 form a biochemical complex within the chromatin fraction (95) and are recruited to damage sites even before the repair factors Rad50 and Rad51 (66), which appear to form a complex together with 53BP1 (65,66). Knocking down of BRCA1 by antisense morpholino oligonucleotides substantially reduced H2AX ubiquitination while increasing its phosphorylation (95). It has therefore been suggested that BRCA1 is part of a late phase of DNA repair, and one function of it is to attenuate and eventually terminate the early response marked by γH2AX (95).
As BRCA1 is detectable by immunoblotting and IIF (66,95), it may easily be included in a marker panel for DLBCL, too (Table 1).
STAT3 and its importance in cancer monitoring
The signal transducer and activator of transcription 3 (STAT3) is a transcription factor regulating genes involved in proliferation, angiogenesis and suppression of apoptosis as well as cell-cycle control, the latter by regulating expression of Myc and cyclin D1 (97). These properties give it the potential to become a strong oncogene if mutated or dysregulated (98). In DLBCL, STAT3 was demonstrated to promote apoptotic resistance in the aggressive ABC subtype (99). Higher levels of STAT3 mRNA were found in ABC compared to GCB DLBCL (105), and high nuclear STAT3 expression in DLBCL tissue samples was associated with poor overall survival (P=0.005) (106). However, not all ABC-type DLBCLs displayed high STAT3 mRNA.
This led to the following conclusions:
- Molecularly different subsets exist within ABC-type DLBCL;
- Differences between STAT3-high and STAT3-low ABC subtypes were suggested to be dependent on expression of anti-apoptotic BCL family members
- STAT3 high cases were BCL-2 low/MCL1;
- STAT3 low cases were BCL-2 high/MCL1 (105).
- STAT3 is associated with Myc and BCL-2 expression;
- Phosphorylated STAT3 levels correlated with poor outcome in DLBCL patients (107).
DLBCL patients positive for pSTAT3 showed a 5-year overall survival of 39.9%, compared to 56.2% in pSTAT3-negative patients (P=0.016) (107). Phosphorylation of tyrosine 705 (pY-STAT3) as observed in the majority led to a significantly better outcome after high dose chemotherapy and autologous stem cell transplantation than patients without this phosphorylated variant with a 5-year event free survival of 92.9% (95% CI: 79.4–100%) versus 47.4% (95% CI: 24.5–70.4%, log-rank P=0.006). A 5-year OS rate of 100% was detected for patients positive for pY-STAT3 compared to an OS rate of 57.9% in patients without tyrosine phosphorylation (P=0.007). Moreover, DLBCL patients harbouring no pY-STAT3, but additionally were positive for p53 expression indicated the worst 5-year OS rate of 40% (P=0.004) (108).
An association between STAT3 and γH2AX was identified in STAT3 knock-out mouse embryonic fibroblasts. When these were treated with etoposide, H2AX was still phosphorylated, but at reduced levels in the STAT3 knock-out compared to normal cells. The reduced formation of γH2AX correlated with ATM activity, and STAT3 appears likely to be important for efficient DNA DSB repair mediated through ATM and γH2AX (109). This makes STAT3 another potential candidate biomarker to predict therapy outcome in DLBCL.
8-Hydroxy-2’-desoxyguanosine
8-Hydroxy-2’-desoxyguanosine (8-OHdG) is a reaction product of DNA oxygenation, which in turn is strongly associated with DNA damage and cancer (110,111). Morning urine concentration of 8-OHdG is used as a biomarker for oxidative stress. Experiments on rats have shown that both 8-OHdG level and frequency of DNA damage increase during physiological aging (112). Quantifying 8-OHdG, however, is not trivial. Techniques such as single-cell gel electrophoresis or comet assay are still used (113), but most often its concentration in liquid samples is quantified by high pressure liquid chromatography (HPLC) (36). Recently, a cell-based automated immunofluorescence method has been established that detected foci after treatment of cells with menadione and H2O2, promising higher efficacy than previous approaches (115). Antibody labelling techniques such as ELISA or IHC are used to detect 8-OHdG. Each method has its advantages and disadvantages and also the type of sample (blood, urine, plasma) should be considered in the detection of 8-OHdG, which has already been discussed elsewhere (116).
Clinically, increased urinary 8-OHdG has been shown in acute myeloid leukaemia patients during chemotherapy (P<0.05), but also in adult T cell leukaemia/lymphoma compared to healthy controls (P<0.05) (117). Similar results could be obtained in patients with prostate or bladder cancer compared to healthy donors with significant increased urinary DNA lesions (bladder cancer P<0.001; prostate cancer P=0.021) (118). These observations suggest that 8-OHdG might be a predictive marker in DLBCL, too (Table 1). Indeed, one study observed a higher 3-year progression-free survival rate in patients with low compared to high 8-OHdG levels (P=0.032, progression-free survival 77.8% vs. 28.6%) (120).
Rad51, a connection between NHEJ and HRe repair
The two major pathways of DNA DSB repair, NHEJ and HRe, are essential for maintaining genomic integrity. Within these two, NHEJ appears to be triggered rather by γH2AX, while HRe is mediated by Rad51, a recombinase critically involved in searching for the intact homologous DNA duplex, DNA pairing and strand exchange.
Rad51 is a homologue of the Escherichia coli RecA and a target for factors such as p53, BRCA1 and BRCA2 which mediate DNA repair, transcription, replication and cell-cycle progression (124,126). Its baseline levels are low and increase markedly upon ionizing irradiation (127,165). This up-regulation depends primarily on a functional HRe repair pathway and Rad53-dependent DNA-damage signalling, less so on the primary DNA damage response as indicated by H2AX activation (124). During DNA DSB introduction, Rad51 foci can be detected at damage sites by immunofluorescence microscopy. Cell lines not capable of HRe due to lack of BRCA1 or BRCA2 are unable to generate Rad51 foci (127). In experiments on mesenchymal stem cells, irradiation induced Rad51 foci only in Ki67-positive, but not in Ki67-negative cells, thus distinguishing proliferating from quiescent cells. Foci formation of γH2AX, by contrast, were induced by radiation in a dose-dependent manner irrespective of proliferation or quiescence (128). In IMR90 human lung fibroblast cells, Rad51 and γH2AX foci co-localized upon irradiation within two to six hours, suggesting that the HRe and NHEJ pathways are co-ordinated and their repair proteins are recruited to specific chromatin sections (66). Furthermore, a positive correlation of Rad51 and γH2AX was observed in endometrial cancer (R =0.318; P=0.001). In 27% of these cancers, loss of nuclear Rad51 was described with γH2AX foci still detectable, suggesting a defect in HRe but not NHEJ (129,165).
Clinically, low Rad51 levels in patients with sporadic primary breast cancer were associated with high histological grade (P=0.031) and high baseline Ki67, a marker for proliferation (P=0.005). In the same study, complete responses were observed in 33% of cancers with low Rad51 versus 3% in the comparison group (P=0.011), emphasizing the dual role of genetic instability for tumorigenesis and for cytotoxic sensitivity (127).
In summary, these results make Rad51 a biomarker for the HRe repair system and a candidate prognostic biomarker in human cancer in general (127-130). In vitro Rad51 inhibition in DT40 chicken B lymphocytes led to cell cycle arrest in G2/S phase and subsequent cell death (134), suggesting a vague relationship between Rad51 and B-cell lymphomas. In the context of lymphoma, however, Rad51 has not yet been addressed clinically (Table 1).
PA28γ and p53 as a link to apoptosis and repair pathways
Alterations in TP53 (p53 gene) were found in many tumours. TP53 mutations are present in approximately 21% of all DLBCL patients, and despite differences in the exact figures among studies, it is associated with poorer survival (135-137). In a small sample of 23 DLBCL patients, Young et al. found a trend towards shorter survival in the presence of TP53 mutations for patients with GCB, but not with ABC subtype (P=0.057 and 0.96, respectively) (137). As both subtypes carry TP53 mutations equally often, this result was to be questioned, and looking at close to 400 patients, Xu-Monette et al. confirmed a significantly worse prognosis in TP53-mutated versus wild-type patients. This applied to both the GCB and ABC subtype with hazard ratios of 0.46 and 0.52, respectively, for overall survival and of 0.45 and 0.57, respectively, for progression-free survival (136).
The most hazardous mutations occur in the DNA-binding domain of TP53 encoded by exon 4 to 8, which is important for the transcription-dependent und transcription-independent activity of p53 protein: 42% of patients carrying a mutation in this region die within one year as opposed to 18% of those with other mutations (136,137).
As mutant p53 variants have a longer half-life than the wild type, apparent overexpression—or rather: increased detection—of p53 protein is due to TP53 mutations and thus to functionally altered p53 protein in most cases (138,139), which may interfere with physiological overexpression and hamper its effect in response to DNA damage and other stressors. In addition to its own direct effects, mutated p53 enhances the effect of Myc rearrangements (138).
To maintain its normal equilibrium, p53 is continuously bound by Mdm2, transported to the cytosol and ubiquitinated for degradation in the proteasome. The proteasome activator PA28γ enhances this process, further reducing the p53 level (140). PA28γ serum levels can be detected by a sandwich ELISA (141), and overexpression of PA28γ was found in various tumours such as colon (142), breast (143), liver (36) and thyroid cancers (144), which makes it likely to be involved in a mechanism of carcinogenesis.
Its role appears to be more complex, though: Live cell imaging has shown that 20S proteasomes locate on the site of DSB, which depends on ATM and PA28γ (145). In this process, the proteasome activator is bound and depleted by ATM (146), and this in turn enhances the accumulation of DNA repair proteins such as MDC1, BRCA1 and 53BP1. Interestingly, in this process γH2AX was only found in later stages after silencing of PA28γ. Thus it is required for the temporal DSB induction (145).
Through its signal in various cancers and its role in DNA repair, PA28γ has emerged as a candidate prognostic biomarker for neoplastic diseases. However, it has not yet been reported on in the context of lymphoma.
Conclusions
The genetic heterogeneity of malignant disease, including aggressive B-Non-Hodgkin Lymphomas such as DLBCL, leads to highly variable results of cytotoxic therapies, ranging from outright resistance to cure. The term “personalized medicine” is mostly used in the context of molecularly targeted substances requiring for their action the presence of a specific altered gene product. However, there is a number of potential biomarkers for monitoring DNA damage and thereby possibly predicting the efficacy of “classical” cytotoxic therapy. In the future this may lead to personalized application of conventional chemotherapy, too, which still is a mainstay of therapy in leukaemia, lymphomas and cancers.
A selection of potential biomarkers has been compiled in this review to shed light on their biological role and potential use as a marker, always with a view on the analytical systems necessary for their detection and quantification.
High levels of the DNA DSB marker γH2AX were found in different tumour types. This biomarker is suitable for the early detection of cancer development because patients with oncological diseases have higher γH2AX foci than healthy volunteers (7,33,48-50,55) and γH2AX has already been investigated in the context of DLBCL (68). From theoretical and practical points of view, it appears to be an attractive candidate biomarker, not least because there are established methods available for the computerized analysis of γH2AX foci in PBMC (51,57,73,75,164,166-170,191). Nevertheless, γH2AX does not provide fully accurate information about the repair of this DNA damage. For this reason, further biomarkers are needed for a study to obtain an exact overview of the course of cancer and DLBCL in particular.
The broad substrate range of ATM (33,48,83) is a relevant example for the close connection of cell-cycle regulation, DNA repair, and apoptosis with cancer. Mutations in the ATM gene also promote DLBCL and complete loss leads to genomic instability and increased sensitivity to radiation (85-87). Like ATM, ATR phosphorylates the histone variant 2AX and is associated with increased radiation sensitivity. Both can be detected by immunostaining (103) and operate in the same signalling pathway (65,131,132,149). They can contribute to the development of tumours in case of alterations. In addition, DNA-PK is also involved in the activation of H2AX and maintains a low level of γH2AX, even if the kinase ATR is knocked out (78). ATR also activates p53 and phosphorylates BRCA1 (147,148). The three kinases are thereby involved in the recognition and repair of DNA damage within a complex network.
BRCA1 is associated with genotoxic stress and γ-radiation and increased γH2AX values could be detected in BRCA1 mutant breast cancer cells (69,95,96). The biomarker is primarily associated with breast and ovarian cancer, but by being associated with γH2AX (69), Rad51 and 53BP1 (65,66), involvement in DLBCL is also likely.
The DNA damage control point protein 53BP1 is involved in DNA repair and copy loss in the 53BP1 locus counts as a predisposing event in DLBCL (157). A correlation of 53BP1 with γH2AX and BRCA1 could be found in breast cancer (69). It appears likely that all three also exert an influence on DLBCL. Complex formation of 53BP1 with Rad50 and Rad51 could also be found. The latter is involved in HRe and also a target for p53 and BRCA1 (124,126). It mediates DNA repair, cell cycle progression and replication and is particularly strongly induced by radiation. In addition, a positive correlation with γH2AX is known in endometrial cancer (129,165). A relationship between Rad51 and B-cell lymphoma could only be found in DT40 chicken cells (134). However, an association is likely and as a marker for HRe, Rad51 would complete an overview of DNA repair. Since oxidative stress can have a major impact on cancer development, 8-OHdG is also of relevance and strongly associated with cancer and DNA damage (110,111). Increased levels could be observed in lymphoma, prostate and bladder cancer (117,118) and a lower 8-OHdG level was associated with longer progression-free survival in DLBCL patients (120).
Overcoming the apoptotic signalling pathway is an important step in promoting cancer progression. Most cancers have elevated levels of Myc, which can lead to overexpression of BCL-2 and thus promote anti-apoptosis in cancer. These two are therefore key players in tumour development and are of predictive value in DLBCL, in particular when both are affected (“double-hit”) (104,185,186). Another apoptosis suppressor and regulator of proliferation is STAT3, which is also involved in the regulation of Myc (97). STAT3 is considered a strong oncogene when misdirected and supports apoptosis resistance in ABC subtypes of DLBCL (105,106). Here the prognosis seems to depend on the phosphorylation pattern, since pSTAT3 carries a poor 5-year OS as opposed to pYSTAT3 which goes along with better survival. A mutation in the p53 gene TP53 is present in 21% of DLBCL patients (135-137), which alters the function of p53 and can therefore also enhance the re-arrangement of Myc (138). p53 is degraded by proteases, including PA28γ (140). Overexpression of this protein has been detected in various tumours (36,142-144). However, it seems to have a far more complex role than has been observed so far, since 20S also localise proteasomes at DSB, a mechanism that depends on PA28γ and ATR and enhances BRCA1 and 53BP1 (145,146).
Most of the biomarkers described here can already be detected by IIF, FISH or IHC and are summarized in Table 1. A combination or multiplex method to check the course of the disease and its development in DLBCL patients would be possible. We were able to show that many biomarkers exert mutual influence on each other and thus a broad spectrum would be required to provide complete and rapid early detection of oncological diseases.
Acknowledgments
Funding: This work has in part been funded by the “Gesundheitscampus Brandenburg - Konsequenzen der alterasassoziierten Zell - und Organfunktionen” initiative of the Brandenburgian Ministry of Science, Research and Culture (MWFK) and the InnoProfile-Transfer 03 IPT 611X project (Federal Ministry of Education and Research).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dirk Roggenbuck) for the series “DNA Damage Assessment for Precision Medicine” published in Journal of Laboratory and Precision Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2019.01.01). The series “DNA Damage Assessment for Precision Medicine” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- WHO. WHO | Cancer [Internet]. 2018 [cited 2018 Apr 13]. Available online: https://web.archive.org/web/20180407122155/http://www.who.int/mediacentre/factsheets/fs297/en/
- Sarkozy C, Coiffier B. Diffuse Large B-cell Lymphoma in the Elderly: A Review of Potential Difficulties. Clin Cancer Res 2013;19:1660-9. [Crossref] [PubMed]
- Habermann TM, Weller EA, Morrison VA, et al. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J Clin Oncol 2006;24:3121-7. [Crossref] [PubMed]
- Tomita N, Takasaki H, Fujisawa S, et al. Standard R-CHOP Therapy in Follicular Lymphoma and Diffuse Large B-Cell Lymphoma. J Clin Exp Hematop 2013;53:121-5. [Crossref] [PubMed]
- Ristow M, Schmeisser K. Mitohormesis: Promoting Health and Lifespan by Increased Levels of Reactive Oxygen Species (ROS). Dose-Response 2014;12:288-341. [Crossref] [PubMed]
- Martinez-Fernandez L, Banyasz A, Esposito L, et al. UV-induced damage to DNA: effect of cytosine methylation on pyrimidine dimerization. Signal Transduct Target Ther 2017;2:17021. [Crossref] [PubMed]
- Bonner WM, Redon CE, Dickey JS, et al. γH2AX and cancer. Nat Rev Cancer 2008;8:957-67. [Crossref] [PubMed]
- Mah LJ, El-Osta A, Karagiannis TC. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 2010;24:679-86. [Crossref] [PubMed]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220-8. [Crossref] [PubMed]
- Hoeijmakers JHJ. DNA Damage, Aging, and Cancer. N Engl J Med 2009;361:1475-85. [Crossref] [PubMed]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature 2000;408:307-10. [Crossref] [PubMed]
- Saha GB, MacIntyre WJ, Go RT. Radiopharmaceuticals for brain imaging. Semin Nucl Med 1994;24:324-49. [Crossref] [PubMed]
- Schiffman JD, Fisher PG, Gibbs P. Early detection of cancer: past, present, and future. Am Soc Clin Oncol Educ Book 2015;57-65. [Crossref] [PubMed]
- Garcia-Villa A, Balsubramanian P, Miller B, et al. Assessment of γ-H2AX levels in circulating tumor cells from patients receiving chemotherapy. Front Oncol 2012;2:128. [Crossref] [PubMed]
- Rack B, Schindlbeck C, Jückstock J, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst 2014;106. [PubMed]
- Symonds EL, Fraser RJL, Young GP. FIT for purpose: enhanced applications for faecal immunochemical tests. J Lab Precis Med 2018;3:28. [Crossref]
- Bulfoni M, Beltrami AP, Cesselli D. Gene expression signatures in circulating tumor cells correlate with response to therapy in metastatic breast cancer. J Lab Precis Med 2018;3:33. [Crossref]
- Zhang Z. The role of big-data in clinical studies in laboratory medicine. J Lab Precis Med 2017;2:34. [Crossref]
- Brower V. Biomarkers: Portents of malignancy. Nature 2011;471:S19-21. [Crossref] [PubMed]
- Sant M, Allemani C, Tereanu C, et al. Incidence of hematologic malignancies in Europe by morphologic subtype: results of the HAEMACARE project. Blood 2010;116:3724-34. [Crossref] [PubMed]
- Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology 2018;50:74-87. [Crossref] [PubMed]
- Swerdlow S, Campo E, Harris N, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Vol. Revised 4th Edition. 2017.
- Coiffier B, Lepage E, Briere J, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med 2002;346:235-42. [Crossref] [PubMed]
- Pfreundschuh M, Trümper L, Osterborg A, et al. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. Lancet Oncol 2006;7:379-91. [Crossref] [PubMed]
- Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103-13. [Crossref] [PubMed]
- Avivi I, Zilberlicht A, Dann EJ, et al. Strikingly high false positivity of surveillance FDG-PET/CT scanning among patients with diffuse large cell lymphoma in the rituximab era. Am J Hematol 2013;88:400-5. [Crossref] [PubMed]
- Camus V, Jardin F, Tilly H. The value of liquid biopsy in diagnosis and monitoring of diffuse large b-cell lymphoma: recent developments and future potential. Expert Rev Mol Diagn 2017;17:557-66. [Crossref] [PubMed]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071-8. [Crossref] [PubMed]
- Poehlmann A, Roessner A. Importance of DNA damage checkpoints in the pathogenesis of human cancers. Pathol Res Pract. 2010;206:591-601. [Crossref] [PubMed]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003;426:194-8. [Crossref] [PubMed]
- Zhou W, Yuan J. Necroptosis in health and diseases. Semin Cell Dev Biol 2014;35:14-23. [Crossref] [PubMed]
- Krysko DV, Vanden Berghe T, D’Herde K, et al. Apoptosis and necrosis: Detection, discrimination and phagocytosis. Methods 2008;44:205-21. [Crossref] [PubMed]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001;27:247-54. [Crossref] [PubMed]
- Cabelof DC, Raffoul JJ, Yanamadala S, et al. Induction of DNA polymerase β-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis 2002;23:1419-25. [Crossref] [PubMed]
- Hung RJ, Hall J, Brennan P, et al. Genetic Polymorphisms in the Base Excision Repair Pathway and Cancer Risk: A HuGE Review. Am J Epidemiol 2005;162:925-42. [Crossref] [PubMed]
- Kondo S, Toyokuni S, Tanaka T, et al. Overexpression of the hOGG1 Gene and High 8-Hydroxy-2′-deoxyguanosine (8-OHdG) Lyase Activity in Human Colorectal Carcinoma: Regulation Mechanism of the 8-OHdG Level in DNA. Clin Cancer Res 2000;6:1394-400. [PubMed]
- Tagesson C, Källberg M, Klintenberg C, et al. Determination of urinary 8-hydroxydeoxyguanosine by automated coupled-column high performance liquid chromatography: a powerful technique for assaying in vivo oxidative DNA damage in cancer patients. Eur J Cancer 1995;31A:934-40. [Crossref] [PubMed]
- Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol 2002;3:21-9. [Crossref] [PubMed]
- Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet 2001;43:71-102. [Crossref] [PubMed]
- Kraemer KH, Lee MM, Andrews AD, et al. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol 1994;130:1018-21. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011;474:609-15. [Crossref] [PubMed]
- Alsop K, Fereday S, Meldrum C, et al. BRCA Mutation Frequency and Patterns of Treatment Response in BRCA Mutation–Positive Women With Ovarian Cancer: A Report From the Australian Ovarian Cancer Study Group. J Clin Oncol 2012;30:2654-63. [Crossref] [PubMed]
- Konstantinopoulos PA, Ceccaldi R, Shapiro GI, et al. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov 2015;5:1137-54. [Crossref] [PubMed]
- Morrison AJ, Highland J, Krogan NJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004;119:767-75. [Crossref] [PubMed]
- Collis SJ, DeWeese TL, Jeggo PA, et al. The life and death of DNA-PK. Oncogene 2005;24:949-61. [Crossref] [PubMed]
- Lee SE, Mitchell RA, Cheng A, et al. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol Cell Biol 1997;17:1425-33. [Crossref] [PubMed]
- Takata M, Sasaki MS, Sonoda E, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 1998;17:5497-508. [Crossref] [PubMed]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687-96. [Crossref] [PubMed]
- Karagiannis TC, El-Osta A. DNA damage repair and transcription. Cell Mol Life Sci 2004;61:2137-47. [Crossref] [PubMed]
- McKinnon PJ, Caldecott KW. DNA Strand Break Repair and Human Genetic Disease. Annu Rev Genomics Hum Genet 2007;8:37-55. [Crossref] [PubMed]
- Reddig A, Lorenz S, Hiemann R, et al. Assessment of modulated cytostatic drug resistance by automated γH2AX analysis. Cytometry A 2015;87:724-32. [Crossref] [PubMed]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene 2002;21:8949-56. [Crossref] [PubMed]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 2000;405:697-700. [Crossref] [PubMed]
- Podhorecka M, Skladanowski A, Bozko P. H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J Nucleic Acids 2010;2010:920161 [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Sordet O, et al. γ-H2AX detection in peripheral blood lymphocytes, splenocytes, bone marrow, xenografts, and skin. Methods Mol Biol 2011;682:249-70. [Crossref] [PubMed]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998;273:5858-68. [Crossref] [PubMed]
- Willitzki A, Lorenz S, Hiemann R, et al. Fully automated analysis of chemically induced γH2AX foci in human peripheral blood mononuclear cells by indirect immunofluorescence. Cytometry A 2013;83:1017-26. [Crossref] [PubMed]
- Redon CE, Nakamura AJ, Gouliaeva K, et al. The Use of Gamma-H2AX as a Biodosimeter for Total-Body Radiation Exposure in Non-Human Primates. PLoS One 2010;5:e15544 [Crossref] [PubMed]
- Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A 2003;100:5057-62. [Crossref] [PubMed]
- Malik A, Kanneganti TD. Function and regulation of IL-1α in inflammatory diseases and cancer. Immunol Rev 2018;281:124-37. [Crossref] [PubMed]
- Marková E, Schultz N, Belyaev DIY. Kinetics and dose-response of residual 53BP1/γ-H2AX foci: Co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol 2007;83:319-29. [Crossref] [PubMed]
- Rappold I, Iwabuchi K, Date T, et al. Tumor Suppressor P53 Binding Protein 1 (53bp1) Is Involved in DNA Damage–Signaling Pathways. J Cell Biol 2001;153:613-20. [Crossref] [PubMed]
- Rasche L, Heiserich L, Behrens JR, et al. Analysis of Lymphocytic DNA Damage in Early Multiple Sclerosis by Automated Gamma-H2AX and 53BP1 Foci Detection: A Case Control Study. PloS One 2016;11:e0147968 [Crossref] [PubMed]
- Rödiger S, Liefold M, Ruhe M, et al. Quantification of DNA double-strand breaks in peripheral blood mononuclear cells from healthy donors exposed to bendamustine by an automated γH2AX assay—an exploratory study. J Lab Precis Med 2018;3:47. [Crossref]
- Ward IM, Chen J. Histone H2AX Is Phosphorylated in an ATR-dependent Manner in Response to Replicational Stress. J Biol Chem 2001;276:47759-62. [Crossref] [PubMed]
- Paull TT, Rogakou EP, Yamazaki V, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000;10:886-95. [Crossref] [PubMed]
- Ivashkevich A, Redon CE, Nakamura AJ, et al. Use of the γ-H2AX Assay to monitor DNA damage and repair in Translational Cancer Research. Cancer Lett 2012;327:123-33. [Crossref] [PubMed]
- Derenzini E, Agostinelli C, Imbrogno E, et al. Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma. Oncotarget 2015;6:6553-69. [Crossref] [PubMed]
- Nagelkerke A, Kuijk SJA, van , Sweep FCGJ, et al. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother Oncol 2011;101:39-45. [Crossref] [PubMed]
- Pang B, Qiao X, Janssen L, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun 2013;4:1908. [Crossref] [PubMed]
- Jain V, Hassan PA, Das B. Radiation-induced conformational changes in chromatin structure in resting human peripheral blood mononuclear cells. Int J Radiat Biol 2014;90:1143-51. [Crossref] [PubMed]
- Rothkamm K, Balroop S, Shekhdar J, et al. Leukocyte DNA Damage after Multi–Detector Row CT: A Quantitative Biomarker of Low-Level Radiation Exposure. Radiology 2007;242:244-51. [Crossref] [PubMed]
- Vandevoorde C, Gomolka M, Roessler U, et al. EPI-CT: in vitro assessment of the applicability of the γ-H2AX-foci assay as cellular biomarker for exposure in a multicentre study of children in diagnostic radiology. Int J Radiat Biol 2015;91:653-63. [Crossref] [PubMed]
- Zhang J, He Y, Shen X, et al. γ-H2AX responds to DNA damage induced by long-term exposure to combined low-dose-rate neutron and γ-ray radiation. Mutat Res Genet Toxicol Environ Mutagen 2016;795:36-40. [Crossref] [PubMed]
- González JE, Lee M, Barquinero JF, et al. Quantitative image analysis of gamma-H2AX foci induced by ionizing radiation applying open source programs. Anal Quant Cytol Histol 2012;34:66-71. [PubMed]
- Lippi G, Buonocore R, Tarperi C, et al. DNA injury is acutely enhanced in response to increasing bulks of aerobic physical exercise. Clin Chim Acta 2016;460:146-51. [Crossref] [PubMed]
- Rassamegevanon T, Löck S, Baumann M, et al. Heterogeneity of γH2AX Foci Increases in Ex Vivo Biopsies Relative to In Vivo Tumors. Int J Mol Sci 2018;19:2616. [Crossref] [PubMed]
- Burma S, Chen BP, Murphy M, et al. ATM Phosphorylates Histone H2AX in Response to DNA Double-strand Breaks. J Biol Chem 2001;276:42462-7. [Crossref] [PubMed]
- Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007;316:1160-6. [Crossref] [PubMed]
- Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A 1980;77:7315-7. [Crossref] [PubMed]
- Savitsky K, Bar-Shira A, Gilad S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 1995;268:1749-53. [Crossref] [PubMed]
- Sun Y, Xu Y, Roy K, et al. DNA Damage-Induced Acetylation of Lysine 3016 of ATM Activates ATM Kinase Activity. Mol Cell Biol 2007;27:8502-9. [Crossref] [PubMed]
- Rotman G, Shiloh Y. ATM: A mediator of multiple responses to genotoxic stress. Oncogene 1999;18:6135-44. [Crossref] [PubMed]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci 2006;31:402-10. [Crossref] [PubMed]
- Grønbaek K, Worm J, Ralfkiaer E, et al. ATM mutations are associated with inactivation of theARF-TP53 tumor suppressor pathway in diffuse large B-cell lymphoma. Blood 2002;100:1430-7. [Crossref] [PubMed]
- Taylor AMR, Harnden DG, Arlett CF, et al. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature 1975;258:427-9. [Crossref] [PubMed]
- Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 2015;149:124-38. [Crossref] [PubMed]
- Price BD, Youmell MB. The Phosphatidylinositol 3-Kinase Inhibitor Wortmannin Sensitizes Murine Fibroblasts and Human Tumor Cells to Radiation and Blocks Induction of p53 following DNA Damage. Cancer Res 1996;56:246-50. [PubMed]
- Hickson I, Zhao Y, Richardson CJ, et al. Identification and Characterization of a Novel and Specific Inhibitor of the Ataxia-Telangiectasia Mutated Kinase ATM. Cancer Res 2004;64:9152-9. [Crossref] [PubMed]
- Batey MA, Zhao Y, Kyle S, et al. Preclinical Evaluation of a Novel ATM Inhibitor, KU59403, In Vitro and In Vivo in p53 Functional and Dysfunctional Models of Human Cancer. Mol Cancer Ther 2013;12:959-67. [Crossref] [PubMed]
- Jacot W, Thezenas S, Senal R, et al. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013;13:523. [Crossref] [PubMed]
- Yossepowitch O, Olvera N, Satagopan JM, et al. BRCA1 and BRCA2 Germline Mutations in Lymphoma Patients. Leuk Lymphoma 2003;44:127-31. [Crossref] [PubMed]
- Liu Q, Guntuku S, Cui X-S, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev 2000;14:1448-59. [PubMed]
- Rosell R, Perez-Roca L, Sanchez JJ, et al. Customized Treatment in Non-Small-Cell Lung Cancer Based on EGFR Mutations and BRCA1 mRNA Expression. PLoS One 2009;4:e5133 [Crossref] [PubMed]
- Krum SA, Dalugdugan E de la R, Miranda-Carboni GA, et al. BRCA1 Forms a Functional Complex with -H2AX as a Late Response to Genotoxic Stress. J Nucleic Acids 2010;2010:801594 [Crossref] [PubMed]
- Scully R, Chen J, Ochs RL, et al. Dynamic Changes of BRCA1 Subnuclear Location and Phosphorylation State Are Initiated by DNA Damage. Cell 1997;90:425-35. [Crossref] [PubMed]
- Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene 2007;26:6724-37. [Crossref] [PubMed]
- Alvarez JV, Frank DA. Genome-wide analysis of STAT target genes: elucidating the mechanism of STAT-mediated oncogenesis. Cancer Biol Ther 2004;3:1045-50. [Crossref] [PubMed]
- Hardee J, Ouyang Z, Zhang Y, et al. STAT3 Targets Suggest Mechanisms of Aggressive Tumorigenesis in Diffuse Large B-Cell Lymphoma. G3 (Bethesda) 2013;3:2173-85. [Crossref] [PubMed]
- Chun HH, Sun X, Nahas SA, et al. Improved diagnostic testing for ataxia–telangiectasia by immunoblotting of nuclear lysates for ATM protein expression. Mol Genet Metab 2003;80:437-43. [Crossref] [PubMed]
- Andegeko Y, Moyal L, Mittelman L, et al. Nuclear Retention of ATM at Sites of DNA Double Strand Breaks. J Biol Chem 2001;276:38224-30. [PubMed]
- Dong J, Zhang T, Ren Y, et al. Inhibiting DNA-PKcs in a non-homologous end-joining pathway in response to DNA double-strand breaks. Oncotarget 2017;8:22662-73. [PubMed]
- Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006;8:37-45. [Crossref] [PubMed]
- Johnson NA, Slack GW, Savage KJ, et al. Concurrent Expression of MYC and BCL2 in Diffuse Large B-Cell Lymphoma Treated With Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone. J Clin Oncol 2012;30:3452-9. [Crossref] [PubMed]
- Lam LT, Wright G, Davis RE, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-κB pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008;111:3701-13. [Crossref] [PubMed]
- Wu ZL, Song YQ, Shi YF, et al. High nuclear expression of STAT3 is associated with unfavorable prognosis in diffuse large B-cell lymphoma. J Hematol Oncol 2011;4:31. [Crossref] [PubMed]
- Ok CY, Chen J, Xu-Monette ZY, et al. Clinical Implications of Phosphorylated STAT3 Expression in de novo Diffuse Large B-cell Lymphoma. Clin Cancer Res 2014;20:5113-23. [Crossref] [PubMed]
- Stewart DA, Bahlis N, Mansoor A. pY-STAT3 and p53 expression predict outcome for poor prognosis diffuse large B-cell lymphoma treated with high dose chemotherapy and autologous stem cell transplantation. Leuk Lymphoma 2009;50:1276-82. [Crossref] [PubMed]
- Barry SP, Townsend PA, Knight RA, et al. STAT3 modulates the DNA damage response pathway. Int J Exp Pathol 2010;91:506-14. [Crossref] [PubMed]
- Reuter S, Gupta SC, Chaturvedi MM, et al. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med 2010;49:1603-16. [Crossref] [PubMed]
- Ristow M. Oxidative metabolism in cancer growth. Curr Opin Clin Nutr Metab Care 2006;9:339-45. [Crossref] [PubMed]
- Fraga CG, Shigenaga MK, Park JW, et al. Oxidative damage to DNA during aging: 8-hydroxy-2’-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A 1990;87:4533-7. [Crossref] [PubMed]
- Flora SJS. Biomarkers in Toxicology. Boston: Academic Press; 2014. 485–519 p. (Metals).
- Hu L, Ru K, Zhang L, et al. Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. Biomark Res 2014;2:3. [Crossref] [PubMed]
- Debelec-Butuner B, Bostancı A, Heiserich L, et al. Automated Cell-Based Quantitation of 8-OHdG Damage. Methods Mol Biol 2016;1516:299-308. [Crossref] [PubMed]
- Korkmaz KS, Butuner BD, Roggenbuck D. Detection of 8-OHdG as a diagnostic biomarker. J Lab Precis Med 2018;3:95. [Crossref]
- Honda M, Yamada Y, Tomonaga M, et al. Correlation of urinary 8-hydroxy-2′-deoxyguanosine (8-OHdG), a biomarker of oxidative DNA damage, and clinical features of hematological disorders: a pilot study. Leuk Res 2000;24:461-8. [Crossref] [PubMed]
- Chiou CC, Chang PY, Chan EC, et al. Urinary 8-hydroxydeoxyguanosine and its analogs as DNA marker of oxidative stress: development of an ELISA and measurement in both bladder and prostate cancers. Clin Chim Acta 2003;334:87-94. [Crossref] [PubMed]
- Barrans S, Crouch S, Smith A, et al. Rearrangement of MYC Is Associated With Poor Prognosis in Patients With Diffuse Large B-Cell Lymphoma Treated in the Era of Rituximab. J Clin Oncol 2010;28:3360-5. [Crossref] [PubMed]
- Pasanen AK, Kuitunen H, Haapasaari KM, et al. Expression and prognostic evaluation of oxidative stress markers in an immunohistochemical study of B-cell derived lymphomas. Leuk Lymphoma 2012;53:624-31. [Crossref] [PubMed]
- Chisholm KM, Bangs CD, Bacchi CE, et al. Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am J Surg Pathol 2015;39:294-303. [Crossref] [PubMed]
- Green TM, Nielsen O, de Stricker K, et al. High Levels of Nuclear MYC Protein Predict the Presence of MYC Rearrangement in Diffuse Large B-cell Lymphoma. Am J Surg Pathol 2012;36:612. [Crossref] [PubMed]
- Sáez AI, Artiga MJ, Romero C, et al. Development of a Real-Time Reverse Transcription Polymerase Chain Reaction Assay for c-myc Expression That Allows the Identification of a Subset of c-myc+ Diffuse Large B-Cell Lymphoma. Lab Invest 2003;83:143-52. [Crossref] [PubMed]
- Cohen Y, Dardalhon M, Averbeck D. Homologous recombination is essential for RAD51 up-regulation in Saccharomyces cerevisiae following DNA crosslinking damage. Nucleic Acids Res 2002;30:1224-32. [Crossref] [PubMed]
- Brugat T, Nguyen-Khac F, Grelier A, et al. Telomere dysfunction-induced foci arise with the onset of telomeric deletions and complex chromosomal aberrations in resistant chronic lymphocytic leukemia cells. Blood 2010;116:239-49. [Crossref] [PubMed]
- Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci 1998;23:247-51. [Crossref] [PubMed]
- Graeser M, McCarthy A, Lord CJ, et al. A marker of homologous recombination predicts pathological complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 2010;16:6159-68. [Crossref] [PubMed]
- Tsvetkova A, Ozerov IV, Pustovalova M, et al. γH2AX, 53BP1 and Rad51 protein foci changes in mesenchymal stem cells during prolonged X-ray irradiation. Oncotarget 2017;8:64317-29. [Crossref] [PubMed]
- Auguste A, Genestie C, Bruyn MD, et al. Refinement of high-risk endometrial cancer classification using DNA damage response biomarkers: a Trans PORTEC initiative. Mod Pathol 2018;31:1851-61. [Crossref] [PubMed]
- Sak A, Grehl S, Erichsen P, et al. gamma-H2AX foci formation in peripheral blood lymphocytes of tumor patients after local radiotherapy to different sites of the body: Dependence on the dose-distribution, irradiated site and time from start of treatment. Int J Radiat Biol 2007;83:639-52. [Crossref] [PubMed]
- Peng Y, Woods RG, Beamish H, et al. Deficiency in the Catalytic Subunit of DNA-Dependent Protein Kinase Causes Down-Regulation of ATM. Cancer Res 2005;65:1670-7. [Crossref] [PubMed]
- Shrivastav M, Miller CA, De Haro LP, et al. DNA-PKcs and ATM Co-Regulate DNA Double-Strand Break Repair. DNA Repair 2009;8:920-9. [Crossref] [PubMed]
- Charrier J-D, Durrant SJ, Golec JMC, et al. Discovery of Potent and Selective Inhibitors of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Protein Kinase as Potential Anticancer Agents. J Med Chem 2011;54:2320-30. [Crossref] [PubMed]
- Sonoda E, Sasaki MS, Buerstedde JM, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 1998;17:598-608. [Crossref] [PubMed]
- Karube K, Enjuanes A, Dlouhy I, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018;32:675-84. [Crossref] [PubMed]
- Xu-Monette ZY, Wu L, Visco C, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2012;120:3986-96. [Crossref] [PubMed]
- Young KH, Weisenburger DD, Dave BJ, et al. Mutations in the DNA-binding codons of TP53, which are associated with decreased expression of TRAILreceptor-2, predict for poor survival in diffuse large B-cell lymphoma. Blood 2007;110:4396-405. [Crossref] [PubMed]
- Wang XJ, Medeiros LJ, Bueso-Ramos CE, et al. P53 expression correlates with poorer survival and augments the negative prognostic effect of MYC rearrangement, expression or concurrent MYC/BCL2 expression in diffuse large B-cell lymphoma. Mod Pathol 2017;30:194-203. [Crossref] [PubMed]
- Xie Y, Bulbul MA, Ji L, et al. p53 Expression Is a Strong Marker of Inferior Survival in De Novo Diffuse Large B-Cell Lymphoma and May Have Enhanced Negative Effect With MYC CoexpressionA Single Institutional Clinicopathologic Study. Am J Clin Pathol 2014;141:593-604. [Crossref] [PubMed]
- Zhang Z, Zhang R. Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 2008;27:852-64. [Crossref] [PubMed]
- Gruner M, Moncsek A, Rödiger S, et al. Increased proteasome activator 28 gamma (PA28γ) levels are unspecific but correlate with disease activity in rheumatoid arthritis. BMC Musculoskelet Disord 2014;15:414. [Crossref] [PubMed]
- Chen D, Yang X, Huang L, et al. The expression and clinical significance of PA28 γ in colorectal cancer. J Investig Med 2013;61:1192-6. [Crossref] [PubMed]
- Wang X, Tu S, Tan J, et al. REG gamma: a potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med Oncol 2011;28:31-41. [Crossref] [PubMed]
- Okamura T, Taniguchi S-I, Ohkura T, et al. Abnormally high expression of proteasome activator-gamma in thyroid neoplasm. J Clin Endocrinol Metab 2003;88:1374-83. [Crossref] [PubMed]
- Levy-Barda A, Lerenthal Y, Davis AJ, et al. Involvement of the nuclear proteasome activator PA28γ in the cellular response to DNA double-strand breaks. Cell Cycle 2011;10:4300-10. [Crossref] [PubMed]
- Stohwasser R. Proteasome Activator 28γ: Impact on Survival Signaling and Apoptosis. Curr Underst Apoptosis - Program Cell Death. 2018. Available online: https://www.intechopen.com/books/current-understanding-of-apoptosis-programmed-cell-death/proteasome-activator-28-impact-on-survival-signaling-and-apoptosis
- Tibbetts RS, Cortez D, Brumbaugh KM, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 2000;14:2989-3002. [Crossref] [PubMed]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20:1803-15. [Crossref] [PubMed]
- Chan DW, Lees-Miller SP. The DNA-dependent Protein Kinase Is Inactivated by Autophosphorylation of the Catalytic Subunit. J Biol Chem 1996;271:8936-41. [Crossref] [PubMed]
- Chen BPC, Uematsu N, Kobayashi J, et al. Ataxia Telangiectasia Mutated (ATM) Is Essential for DNA-PKcs Phosphorylations at the Thr-2609 Cluster upon DNA Double Strand Break. J Biol Chem 2007;282:6582-7. [Crossref] [PubMed]
- Solier S, Sordet O, Kohn KW, et al. Death Receptor-Induced Activation of the Chk2- and Histone H2AX-Associated DNA Damage Response Pathways. Mol Cell Biol 2009;29:68-82. [Crossref] [PubMed]
- Peasland A, Wang L-Z, Rowling E, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer 2011;105:372-81. [Crossref] [PubMed]
- ClinicalTrails.gov. Identifier NCT02223923, Phase I Study to Assess Safety of AZD6738 Alone and in Combination With Radiotherapy in Patients With Solid Tumours [Internet]. ClinicalTrails.gov. 2014 [cited 2018 Sep 18]. Available online: https://clinicaltrials.gov/ct2/show/NCT02223923
- Rivera-Calzada A, Spagnolo L, Pearl LH, et al. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep 2007;8:56-62. [Crossref] [PubMed]
- Spagnolo L, Rivera-Calzada A, Pearl LH, et al. Three-Dimensional Structure of the Human DNA-PKcs/Ku70/Ku80 Complex Assembled on DNA and Its Implications for DNA DSB Repair. Mol Cell 2006;22:511-9. [Crossref] [PubMed]
- DeFazio LG, Stansel RM, Griffith JD, et al. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 2002;21:3192-200. [Crossref] [PubMed]
- Takeyama K, Monti S, Manis JP, et al. Integrative analysis reveals 53BP1 copy loss and decreased expression in a subset of human diffuse large B-cell lymphomas. Oncogene 2008;27:318-22. [Crossref] [PubMed]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003;5:675-9. [Crossref] [PubMed]
- Gupta A, Hunt CR, Chakraborty S, et al. Role of 53BP1 in the Regulation of DNA Double-Strand Break Repair Pathway Choice. Radiat Res 2014;181:1-8. [Crossref] [PubMed]
- Iwabuchi K, Li B, Massa HF, et al. Stimulation of p53-mediated Transcriptional Activation by the p53-binding Proteins, 53BP1 and 53BP2. J Biol Chem 1998;273:26061-8. [Crossref] [PubMed]
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun 1984;123:291-8. [Crossref] [PubMed]
- Ismail IH, Wadhra TI, Hammarsten O. An optimized method for detecting gamma-H2AX in blood cells reveals a significant interindividual variation in the gamma-H2AX response among humans. Nucleic Acids Res 2007;35:e36 [Crossref] [PubMed]
- Palla VV, Karaolanis G, Katafigiotis I, et al. gamma-H2AX: Can it be established as a classical cancer prognostic factor? Tumour Biol 2017;39:1010428317695931 [Crossref] [PubMed]
- Böcker W, Iliakis G. Computational Methods for Analysis of Foci: Validation for Radiation-Induced γ-H2AX Foci in Human Cells. Radiat Res 2006;165:113-24. [Crossref] [PubMed]
- Brown JAL, Eykelenboom JK, Lowndes NF. Co-mutation of histone H2AX S139A with Y142A rescues Y142A-induced ionising radiation sensitivity. FEBS Open Bio 2012;2:313-7. [Crossref] [PubMed]
- Herbert AD, Carr AM, Hoffmann E. FindFoci: A Focus Detection Algorithm with Automated Parameter Training That Closely Matches Human Assignments, Reduces Human Inconsistencies and Increases Speed of Analysis. PLoS One 2014;9:e114749 [Crossref] [PubMed]
- Hernández L, Terradas M, Martín M, et al. Highly Sensitive Automated Method for DNA Damage Assessment: Gamma-H2AX Foci Counting and Cell Cycle Sorting. Int J Mol Sci 2013;14:15810-26. [Crossref] [PubMed]
- Ivashkevich AN, Martin OA, Smith AJ, et al. γH2AX foci as a measure of DNA damage: A computational approach to automatic analysis. Mutat Res 2011;711:49-60. [Crossref] [PubMed]
- Lapytsko A, Kollarovic G, Ivanova L, et al. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics 2015;16:392. [Crossref] [PubMed]
- Martin OA, Ivashkevich A, Choo S, et al. Statistical analysis of kinetics, distribution and co-localisation of DNA repair foci in irradiated cells: Cell cycle effect and implications for prediction of radiosensitivity. DNA Repair 2013;12:844-55. [Crossref] [PubMed]
- Reddig A, Fatahi M, Friebe B, et al. Analysis of DNA Double-Strand Breaks and Cytotoxicity after 7 Tesla Magnetic Resonance Imaging of Isolated Human Lymphocytes. PLoS One 2015;10:e0132702 [Crossref] [PubMed]
- Sowa M, Reddig A, Schierack P, et al. Phosphorylated histone 2AX foci determination in capillary blood mononuclear cells. J Lab Precis Med 2018;3:45. [Crossref]
- Willitzki A, Hiemann R, Peters V, et al. New Platform Technology for Comprehensive Serological Diagnostics of Autoimmune Diseases. Clin Dev Immunol 2012;2012:284740 [Crossref] [PubMed]
- Reddig A, Roggenbuck D, Reinhold D. Comparison of different immunoassays for γH2AX quantification. J Lab Precis Med 2018;3:80. [Crossref]
- Ruhe M, Dammermann W, Lüth S, et al. Effect of Cryopreservation on the Formation of DNA Double Strand Breaks in human Peripheral Blood Mononuclear Cells. J Cell Biotechnol 2018. [Epub ahead of print].
- Raap AK. Advances in fluorescence in situ hybridization. Mutat Res 1998;400:287-98. [Crossref] [PubMed]
- Nakamura AJ, Redon CE, Bonner WM, et al. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging 2009;1:212-8. [Crossref] [PubMed]
- Franco S, Gostissa M, Zha S, et al. H2AX Prevents DNA Breaks from Progressing to Chromosome Breaks and Translocations. Mol Cell 2006;21:201-14. [Crossref] [PubMed]
- Yin B, Savic V, Juntilla MM, et al. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med 2009;206:2625-39. [Crossref] [PubMed]
- Eischen CM, Packham G, Nip J, et al. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 2001;20:6983-93. [Crossref] [PubMed]
- McMahon SB. MYC and the Control of Apoptosis. Cold Spring Harb Perspect Med 2014;4:a014407 [Crossref] [PubMed]
- Lynnhtun K, Renthawa J, Varikatt W. Detection of MYC rearrangement in high grade B cell lymphomas: correlation of MYC immunohistochemistry and FISH analysis. Pathology 2014;46:211-5. [Crossref] [PubMed]
- Muñoz-Mármol AM, Sanz C, Tapia G, et al. MYC status determination in aggressive B-cell lymphoma: the impact of FISH probe selection. Histopathology 2013;63:418-24. [Crossref] [PubMed]
- Zhou K, Xu D, Cao Y, et al. C-MYC Aberrations as Prognostic Factors in Diffuse Large B-cell Lymphoma: A Meta-Analysis of Epidemiological Studies. PLoS One 2014;9:e95020 [Crossref] [PubMed]
- Trop-Steinberg S, Azar Y. Is Myc an Important Biomarker? Myc Expression in Immune Disorders and Cancer. Am J Med Sci 2018;355:67-75. [Crossref] [PubMed]
- Reyes-González JM, Armaiz-Peña GN, Mangala LS, et al. Targeting c-MYC in platinum-resistant ovarian cancer. Mol Cancer Ther 2015;14:2260-9. [Crossref] [PubMed]
- Klapper W, Kreuz M, Kohler CW, et al. Patient age at diagnosis is associated with the molecular characteristics of diffuse large B-cell lymphoma. Blood 2012;119:1882-7. [Crossref] [PubMed]
- Liu Y, Hernandez AM, Shibata D, et al. BCL2 translocation frequency rises with age in humans. Proc Natl Acad Sci U S A 1994;91:8910-4. [Crossref] [PubMed]
- Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2—associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL). Blood 2003;101:4279-84. [Crossref] [PubMed]
- Thunberg U, Amini R-M, Linderoth J, et al. BCL2 expression in de novo diffuse large B-cell lymphoma partly reflects normal differences in age distribution. Br J Haematol 2009;146:683-4. [Crossref] [PubMed]
- Horn S, Barnard S, Brady D, et al. Combined Analysis of Gamma-H2AX/53BP1 Foci and Caspase Activation in Lymphocyte Subsets Detects Recent and More Remote Radiation Exposures. Radiat Res 2013;180:603-9. [Crossref] [PubMed]
Cite this article as: Ruhe M, Rabe D, Jurischka C, Schröder J, Schierack P, Deckert PM, Rödiger S. Molecular biomarkers of DNA damage in diffuse large-cell lymphoma—a review. J Lab Precis Med 2019;4:5.


