
Open source bioimage informatics tools for the analysis of DNA damage and associated biomarkers
Introduction
Deoxyribonucleic acid (DNA) is exposed to several endogenous as well as exogenous factors that can profoundly change and damage its structure and function (1). DNA damage response (DDR) mechanisms signal the presence of DNA damage and exert their repair by often redundant pathways. Cells counteract DNA damage events by various repair mechanisms like non-homologous end joining (NHEJ), homologous recombination (HR) (2), mismatch repair and base or nucleotide excision repair (3-5). The detection and assessment of DNA damage like DNA double-strand breaks (DSBs) are of interest in clinical diagnostics, cancer research, radiation therapy, chemotherapy, forensics and life sciences (6-10). DSBs are a common type of DNA damage. They are accompanied by the phosphorylation of the histone 2AX (H2AX), which can be detected as discrete spots, the so-called γH2AX focal points (foci), using immunofluorescence detection methods (e.g., fluorescence microscopy) (2,11-13).
The quantification of foci is considered as a tool for precision medicine. Precision medicine uses biomarkers to stratify patients. One goal is to develop a tailor-made therapy for each patient, which can be based on the automated processing of large image data sets and the classification of defined biomarker image patterns (14). Bioimage informatics is a branch of bioinformatics that deals with informatics tools to support the analysis and numerical description of images in biological and biomedical studies.
The analysis of DNA damage is also discussed as a tool for forensics. DNA in environmentally contaminated samples often contains several complex lesions and is highly fragmented (15,16). A positive correlation between the post-mortem interval (PMI) and the strength of DNA fragmentation and DNA damage has been reported. DNA damage can lead to strong enough fragmentation that PCR-based methods will no longer be applicable for analysis, which will lead e.g., to the failure of DNA genotyping (17,18). Previously it was shown that γH2AX foci are formed preferentially in actively transcribing euchromatin following γ-irradiation (19). Thus, also microsatellite DNA is affected, which is used for forensic DNA analysis, as it is widely distributed over euchromatin (20). In this case, DSB analysis with imaging techniques and bioimage informatics may reduce the number of false-negative results. As of completion of this study there was no application of γH2AX foci analysis in forensics.
Manual counting, the traditional method for quantifying foci in microscopic images, is criticized as time-consuming and user-unfriendly. Therefore, software packages were developed to support the foci counting. Some of these software packages, such as FindFoci, were specifically designed for the analysis of DSBs while other software packages such as CellProfiler serve as multipurpose tools also applicable for foci quantification. This review provides a brief overview about DNA damage and open source software for automated analysis of DSBs from image data. The study is aimed at researchers who have no background in bioimage informatics (21-23). The authors of this study hope to support researchers during their quest for appropriate software.
Biomarkers for the analysis of DNA damage
DSBs are common and often result in fatal DNA damage at the cellular level. H2AX phosphorylated at serine 139 (γH2AX) is a surrogate biomarker of DSBs (11,24-26). γH2AX is an early biomarker that serves as signal for the recruitment and accumulation of protein complexes in the close vicinity of DSBs supporting efficient recognition and repair thereof (27). However, γH2AX is also induced by some non-DSB conditions during apoptosis, which may lead to an overestimation of genotoxic agents and thus impairs the accuracy of DSB assessments (28). This problem can be addressed by the analysis of γH2AX recruited mediators and transducers such as the DNA-damage response (DDR) sensor p53-binding protein 1 (53BP1). 53BP1 is co-localized with γH2AX and facilitates the DDR. Similar to γH2AX, 53BP1 accumulates immediately at the damaged DNA sites as foci. Therefore, 53BP1 is often quantified along with γH2AX (29-31). Understanding the temporal sequence of DSB biomarkers during the DDR could help to develop new cancer therapies. In this context, the nucleus size serves as an indicator of cell viability, since cytostatic drugs such as etoposide can cause nuclear expansion (32).
Löbrich et al. discussed pitfalls and use cases of γH2AX assays. Among other things, they underlined the importance of the cell cycle on γH2AX formation during irradiation. To optimize the assay, they proposed to monitor the cell cycle with cell cycle markers such as Cyclin A or CENP-F. Consideration of chromatin remodeling might not only be beneficial for the analysis of DSBs, but could ultimately improve precision medicine by providing detailed phenotypic information (33).
The biomarker Ku70 enables to gauge the cellular DDR capacity and the health status of cells (34,35). Other biomarker proteins, such as 53BP1, Rad50, Rad51, Nbs1 and the product of the tumor suppressor gene BRCA1, are associated with the repair of DSBs and accumulate as foci in the immediate vicinity of DBSs (36,37). Image analysis tools are used for spatiotemporal cell cycle phase analyses (38,39).
Another application of these biomarkers is the concurrent assessment of apoptotic processes, where nuclei and chromatin are condensed. This leads to an accumulation and aggregation of γH2AX as well as translocation of proteins in the plasma membrane (40). Foci forming biomarkers of the DDR can be characterized by measuring total nuclear fluorescence signal, estimating foci positivity and scoring the number of foci per nucleus or per nuclear area (41).
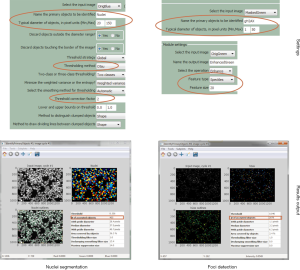
Biomarkers differ in their characteristics and do not necessarily occur as discrete spots (foci). Some occur as homogeneous or cytoplasmic. They may appear as centromeric, nucleolar or speckled pattern or as nuclear dots (42). Biomarkers with dense fine speckled pattern like dense fine speckles 70 kD protein (DFS70) are challenging to quantify (43). Here, the image analysis demands software with robust pattern recognition algorithms. In consequence, software for DSB assessment needs to quantify such parameters and should follow guidelines for the image analysis (Figure 1). An example is the estimation of the Mitotic Index (MI) provided by the CellProfiler open source software. This can be used for the assessment of the cellular viability by automatic detection of mitotic cells and nuclei (44).
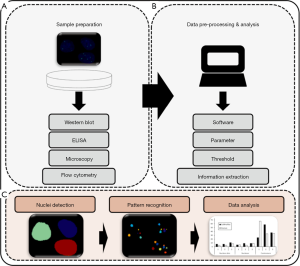
Techniques for assessment of DNA damage
Image analysis is influenced by the experimental conditions like the treatment of the specimen as well as the used microscopic technique (Figure 1). The image acquisition is limited by the optical system and sensors (45). Since this review focuses on the software packages for quantification of foci data, we would like to refer the reader to the references given and the study of Jennifer C. Waters (46).
Automatic foci analysis is limited by the sensitivity of the γH2AX foci labeling and the detection system. Rogakou et al. estimated that approximately up to 2,000 H2AX molecules are phosphorylated per DSB. However, this does not tell how many specific anti-γH2AX antibodies bind to a focus (11). It is assumed that a few hundred of secondary antibody molecules generate a signal that exceeds the threshold.
Relative quantification of DNA damage
Bioanalytical methods for the assessment of DNA damage and DSB quantification have been reviewed extensively (47). The assessment of DNA damage can be carried out by the cytokinesis-blocked micronucleus assay as a standard biodosimetry assay (48), the fluorimetric detection of alkaline DNA unwinding (FADU) assay for detection of DNA strand breaks (49,50) and immunoblot analysis in combination with densitometry to measure the total amount of γH2AX.
Flow cytometry is also used to measure the total intensity of γH2AX fluorescence signals (Figure 1) (51). Most image cytometers work in 2D only and the analysis of the spatial distribution and overlapping foci is demanding. Wadduwage et al. developed an in-house, high-throughput image cytometer with a HiLo wide-field depth resolved imaging and a remote z-scanning technique (52,53). They achieved imaging speeds as high as 800 cells/second with a 3D resolution (54). The fixation with paraformaldehyde, permeabilization with Triton/PBS, blocking with BSA and immunostaining with fluorescent antibodies are similar to standard sample preparation for flow cytometry. One may argue, that the 3D cell structure of the image cytometer or fluorescence microscope approach resemble more closely the native cell structure. However, image cytometers have several limitations. In some cases loss of sample material during the scanning process is unavoidable. Repeated measurements are impossible in this case. They are technically more demanding and their maintenance requires more effort than conventional fluorescence microscopes.
The comet assay enables the detection of single-stranded DNA breaks, variations in the DNA repair pathways (incl. nucleotide excision repair, NHEJ, mismatch repair, base excision repair) besides the analysis of DSBs. The formation of comet-like tails in the electrophoretic field can be readily visualized and subsequently analyzed by image software employing a set of numeric descriptors and scores (55-58). There is still an ongoing discussion if manual or automatic scoring in this context is the best approach (59,60).
All these methods do not report the number of DSBs per cell but a sum signal of all cells analyzed (relative quantification).
Absolute quantification of DNA damage by bioimage informatics
The region of interest (ROI) for foci image analysis is the nucleus. A standard approach to label the nucleus is by chromatin staining substances (e.g., DAPI). Stained regions are analyzed by projection of a maximum intensity algorithm of z-stacks and subsequent deconvolution (61). Others used Deep Learning strategies for nucleus segmentation (62).
Immunofluorescence staining of γH2AX to detect DSBs is frequently used (47). This staining results in discrete fluorescent foci in the nucleus. Foci are countable (absolute quantification) by fluorescence microscopy in combination with digital image analysis. Basic steps for quantification of foci analysis are (I) image capturing, (II) thresholding/segmentation, (binary) mask creation, and (III) foci counting (63).
Thresholding (value that separates between the nucleus and the background) is a critical step. Otsu’s automatic threshold method is widely used (64). Several algorithms are available for image segmentation, shape fitting, and feature extraction. Gray level histograms, fuzzy set theoretic approaches and second-order edge detection and principal component analysis with thresholding have been proposed (65,66). Noise can be dealt with by Markov Random Field (MRF) models and neural network architectures (67,68).
Pixel with a higher intensity than the background pixels can be considered as a focus or part thereof. Further foci parameters like the height, width, shape and distance from other foci can be used to minimize the detection of false-positive foci (63,69).
Depending on the analysis concept, the evaluation process can be carried out in a semi-automatically fashion. Here, manual operations are needed for nuclei detection (61). Thereby, each image is analyzed step-wise or in a batch (24,29,48) or a set of images is automatically processed by the software with user defined parameters (69). There is a tradeoff between the number of required user interaction and the usability. Software packages requiring the user to adjust many parameters for the analysis impact the usability negatively and may result in a counter-intuitive analysis tool (70,71).
Image-based assays with corresponding software have been made commercially available [e.g., Aklides, Medipan, Germany (42); EUROPattern, EUROIMMUN AG, Germany (72); Metafer, Metasystems, Germany (73); NOVA View®, Inova Diagnostics, Inc., USA (74)]. High-content and high-throughput imaging technologies for DNA damage assessment on cellular molecular level allows researchers to better understand of biomedical processes (71).
To avoid problems and to ensure high quality data after image analysis, suitable microscopic techniques are crucial. Studies of multivariate features use mainly fluorescence microscopy in complex biology systems. High-resolution microscopy methods are commonly used in research for the analysis of the spatial distribution and co-localization of DSBs (71). Methods include stimulated emission depletion (STED), ground-state depletion microscopy followed by individual molecule return (GSDIM) and structured illumination microscopy (SIM). Using improved confocal microscopy, Britton et al. reported that Ku70/Ku80 foci are smaller than γH2AX analogues, which indicates that Ku70/Ku80-induced foci would constitute more accurate biomarkers of DSB localization (75).
Confocal laser scanning microscopy is still used extensively and remains a standard approach for foci detection (21). Characteristics of several microscope types are described by Ronneberger et al. (76). In particular, mechanistic questions on the spatiotemporal aspects and the quantification of size or volume and relative three-dimensional distribution of a feature (e.g., foci) within individual nuclei can be investigated (77). Cells as three-dimensional objects have varying nucleus depth, which not only varies between different cell lines but also between individual cells. This has to be taken into account for robust analysis. Different approaches may also be needed for analysis of adherent compared to suspended cells. The latter may require more z-stacks compared to adherent cells. Vasireddy and colleagues demonstrated a method for detection and quantification of γH2AX foci in non-adherent cells, with special focus on the co-localization of γH2AX foci with other epigenetic markers. Their analysis used bioimage software for detection and quantification of foci as well for the creation of three-dimensional foci distribution maps from z-stack images (78).
During the analysis, the influence of the hardware components on the analysis quality must be taken into account. Microscopy experts should be involved to ensure that an optimal image quality is achieved. High magnifications and low aperture values lead to incorrect assignments of focusing to several z-stacks and thus to poor data quality. As a result, the total number of foci in a sample may be overestimated. Confocal laser microscopy is a method that reduces such problems because signals outside a defined z-stack are treated as fuzzy signals. The Nyquist theorem is a method of roughly estimating the number of z-stacks required (79). Standard fluorescence microscopy is still the dominant for foci analysis since confocal microscopes are expensive and thus not ubiquitous available, especially for small research teams.
Cell models to study DNA double-strand breaks
Different human cell lines and types are used as in vitro models to study individual differences and cell type-specific reactions of DDR. For example, peripheral blood mononuclear cells (PBMCs) are commonly used as in vitro model due to the broad availability of these cells in clinical research (80). PBMCs isolated from female patients suffering from high-grade serous ovarian cancer were used to investigate the concurrent induction of γH2AX and MRE11 to ascertain homologous recombination deficiency. In this case, biomarkers were stained with fluorophore-conjugated antibodies and subsequently analyzed by high-throughput flow-cytometry (81). However, the question if PBMCs as in vitro model accurately resemble the dose-effect-relations expected in vivo remains to be answered (9).
The automated analysis of such cell models requires profound knowledge about the cell morphology:
- Variable cell shapes and dynamic morphological features require an individual adoption and validation of pattern recognition algorithms for each cell line. Böcker and Iliakis investigated three cell lines (HeLa, A549 and MRC5) and demonstrated that they exhibited varying nuclear morphology and γH2AX foci size. Thus, HeLa cell nuclei were nearly spherical and smaller in average, while A549 and MRC5 cells had a much flatter nucleus. Conversely, the focus size was larger in HeLa and MRC5 cells than in A549 cells (21). During our analysis we noted that some software discussed in the later sections failed to report meaningful results if this was not adjusted initially.
- Malignant cells have higher DSB base-line levels that may even vary under differing culture conditions. Senescent cells show normal numbers of endogenous γH2AX foci irrespective of origin (82). Studies listed in Table 1 point to cell lines where DSBs were induced.
Table 1
Representative studies for DSB assessment using differing cellular systemsIn vitro model Origin DSB inducer Assessed biomarker Assay technique Type Ref. CaCo2 Intestine (colon) Acetamiprid γH2AX IF, CA m (83) HEK293 Kidney Irradiation γH2AX, 53BP1, MRE11, BRCA1, NFBD1 IF, FACS m (84) HeLa Uterus Irradiation γH2AX, 53BP1 IF m, s (30) HEp-2 Larynx Arecoline γH2AX, ATM, p53, Chk1/2 IF, WB, HCR m (85) HepG2 Liver Etoposide γH2AX FACS s (86) LoVo Intestine (colon) NU7441, Irradiation, Etoposide, Doxorubicin γH2AX FACS s (87) PBMCs Blood (venous) Etoposide γH2AX IF, WB a (69) Irradiation γH2AX, 53BP1 IF m (88) DSB, double-strand break; a, automatic; CA, comet assay; HCR, host cell reactivation assay; IF, immunofluorescence; m, manual (=optical); PBMCs, peripheral blood mononuclear cells; s, semi-automatic; WB, western blot.
Open source software packages for immunofluorescence pattern evaluation and foci quantification
For this review, peer-reviewed open source software packages were considered to provide an overview of ready-to-use software for automated immunofluorescence pattern evaluation and quantification of DNA damage responses (e.g., DSBs). Our literature research showed that ImageJ, CellProfiler, Icy, FociCounter and the ImageJ plug-in FindFoci are among the most frequently cited open source software in the year 2018. They are designed to process high quality images or z-stacks obtained by either high-end confocal laser scanning microscopy or wide-angle fluorescence microscopy. The latter usually have an increased background signal due to a less well-defined focal plane (61).
Open source software packages have several benefits over closed source software such as:
- An accessible source code that can be modified to specific needs;
- A transparent and reproducible result generation process;
- Free availability for the scientific community.
All software is based on a similar workflow (Figure 2) (83). They represent research tools with great potential for diagnostic and research applications and allow to tailor the program to specific tasks (e.g., machine learning) (63,84). Necessary image processing tasks are often performed based on ImageJ (89). Different design approaches are available:
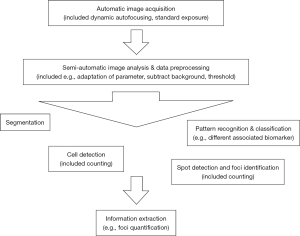
- Macros [e.g., ImageJ (85,86), Focinator (87,88), BIC Macro Toolkit (90)];
- Plug-ins [e.g., Icy (91), ImageJ (85,86)] for available software;
- Pipelines [e.g., CellProfiler (89,92,93)], which are based on the use of freely available or commercial software frameworks such as ImageJ and NIH Image;
- Standalone software like FociCounter (22) or ilastik (94).
Open source digital image analysis software packages
Due to the complexity of the software packages, it is not feasible to discuss all details of the operation. The literature cited in Table 2 points to quantification of results and the usability of the software. While CellProfiler, Icy and ImageJ are able to analyze large image sets and output detailed results (e.g., area, intensity) in different file formats, the standalone software FociCounter is focused on the analysis of single cells.
Table 2
| Software Packages | Program details | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ODP | License | Advanced data exchange/data output | Throughput | Programming language | Usability | Microscopy | Associated biomarker | Ref. | |
| CellProfiler | 10/2006 | GPL v2 | MATLAB (.mat), HDF5 (.h5), EXCEL sheets | Multiple cells simultaneously | Python | Advanced | Fluorescence | Yes | (95) |
| FociCounter | 12/2009 | GNU GPL | – | Restricted to single cells | Python | Novice | All | No | (22) |
| Icy | 06/2012 | GPLv3 | MATLAB (.mat), HDF5 (.h5), EXCEL sheets | Multiple cells simultaneously | Java | Advanced | All | Yes | (96) |
| ImageJ and ImageJ2/Fiji | 06/2012 | PD | Internal data table | Multiple cells simultaneously | Java | Advanced | All | Yes | (89,92,97) |
**, via implemented ImageJ. CF, confocal fluorescent laser scanning microscope; GPL, GNU General Public License; HDF, hierarchical data format; NA, not available; OPD, online publication date; PD, public-domain; WF, wide-field fluorescent microscope.
ImageJ—Java-based image processing program
ImageJ stands out by a stable program interface with the ability for macro- or plug-in based expansion. A commonly used distribution of ImageJ is Fiji (recursive acronym for Fiji Is Just ImageJ), which bundles ImageJ with many useful plug-ins. Repetitive tasks such as multistep image processing via color balance adjustment, histogram equalization, blurring or thresholding can be automated via custom-built macros. An advantage of ImageJ is the availability of numerous plug-ins for a variety of analysis tasks. The plug-ins are generally well documented and are modifiable to adapt them for different analysis needs. Furthermore, it has the ability for batch processing, which allows the analysis of a large amount of images semi-automatically (85,99). To investigate the inner workings of plug-ins and macros, ImageJ also allows a step-by-step analysis. Since it is presumably one of the most used tools for digital image analysis, we show a brief outline for the analysis:
- Digital image pre-processing can be performed to minimize noise by different blurring methods, such as Box or Gaussian blur. However, blurring always results in loss of contrast, which might negatively influence further processing steps.
- Subsequently, the images can be segmented by a threshold to facilitate recognition and classification of different objects (Region of Interests).
- After watershed transformation, the number of detected objects can be determined using the option “Analyze Particles”.
- Furthermore, the determination of foci number can be performed by the “FindMaxima” option using spot intensity (Figure 3).
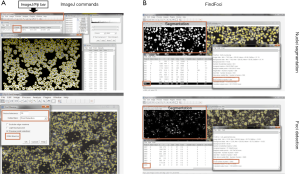 Figure 3 Comparison of implemented ImageJ commands with the ImageJ plug-in FindFoci with respect to nuclei segmentation and foci detection. (A) By usage of following implemented ImageJ commands it is possible to segment and detect objects without plug-ins: Gaussian Blur → Substract Background → Threshold → Watershed → Analyze Particles. As results you will get in addition to the number of counted objects for example also their area and their average size shown at the screenshot top left under the ImageJ/Fiji bar. In contrast, for foci detection only the FindMaxima command is needed, which is suitable for fast detection of punctiform objects by adjusting the parameters shown in the picture bottom left. (B) The ImageJ plug-in FindFoci enables the automated recognition of (punctiform) objects. By the integrated special features FindFoci Optimizer and FindFoci Batch it is possible to analyze numerous images under optimal settings. In addition to the number of segmented objects and focal events, for example the size and the respective position you get. Furthermore, the comparison of both approaches showed that both recognize the objects equally and in the same dimension. The important details are framed in red.
Figure 3 Comparison of implemented ImageJ commands with the ImageJ plug-in FindFoci with respect to nuclei segmentation and foci detection. (A) By usage of following implemented ImageJ commands it is possible to segment and detect objects without plug-ins: Gaussian Blur → Substract Background → Threshold → Watershed → Analyze Particles. As results you will get in addition to the number of counted objects for example also their area and their average size shown at the screenshot top left under the ImageJ/Fiji bar. In contrast, for foci detection only the FindMaxima command is needed, which is suitable for fast detection of punctiform objects by adjusting the parameters shown in the picture bottom left. (B) The ImageJ plug-in FindFoci enables the automated recognition of (punctiform) objects. By the integrated special features FindFoci Optimizer and FindFoci Batch it is possible to analyze numerous images under optimal settings. In addition to the number of segmented objects and focal events, for example the size and the respective position you get. Furthermore, the comparison of both approaches showed that both recognize the objects equally and in the same dimension. The important details are framed in red.
The image processing program ImageJ/Fiji along with its macros and plug-ins has been used to solve many biological questions based on accurate and reproducible image analysis (85). User-friendliness and the availability of macros for automation of standard functions make ImageJ/Fiji a suitable choice for high-throughput image analysis. In many cases, advanced knowledge of programming languages is not required. However, when it comes to high-quality analysis routines (e.g., compensation for inhomogeneous illumination, sharpness) advanced programming skills and deep knowledge of digital image processing is indispensable.
FindFoci, FoCo and Focinator—focus detection algorithms and automated high-throughput foci counting
FindFoci is an ImageJ plug-in for an automated foci recognition, which is based on ImageJ’s FindMaxima operation. Detecting and counting of objects via size and intensity is based on thresholding and optional pre-processing by Gaussian blur. The user-friendly interface, comprehensive documentations and additional plug-ins like the FindFoci Optimizer or FindFoci Batch facilitate reproducible foci quantification. Parameters can be adjusted such as the thresholding method (e.g., Otsu’s method) or the minimal ROI size. The options of the FindFoci plug-in are extensively documented in an online manual (96).
Other plug-ins and macros like Focinator or BIC macro toolkit can also be employed for foci counting (87,90). The developers of Focinator argued that most of the foci counting software available lacks a graphical user interface (GUI) (87). Indeed, this is an obstacle for many users to adopt software in their scientific work process, because the operation of software from the command-line is rather complicated for most users to achieve their goals. But not only pure ImageJ-based programs were designed: Lapytsko et al. developed a simple and robust quantification algorithm for nuclear foci, called FoCo which is a Matlab program using ImageJ for image processing. It can be used even for low signal to noise ratios and densely distributed foci (61).
AutoFoci—automated high-throughput foci detection
Lengert and colleagues implemented an automated focus counting method called AutoFoci to count γH2AX and 53BP1 foci in low-dose irradiated cells (95). AutoFoci is a bioimage informatics tool based on Java/ImageJ for high-throughput analysis of cell images (Figure 4). It records various object properties such as co-localizing γH2AX/53BP1 foci, their intensity, size and sharpness. According to the authors, it is suitable for many biological screening approaches. Depending on the resolution of the cell images, AutoFoci requires user-defined input parameters for reliable automated analysis. For their method, the authors combined a scanning fluorescence microscope with an autofocus function (~50 cells/10×10 fields). Using the µManager software (100), the cells in each field in the blue channel (DAPI staining) were detected and recorded in a z-stack of five images per DNA damage marker. The CellECT software (101) was used to identify individual cell nuclei and generate individual images thereof. The image with the highest contrast was selected from the z-stack for further analysis and foci counting. With this approach the background signals should be reduced in comparison to a maximum intensity projection. In addition, algorithms were implemented to identify all S- and G2-phase cells, potentially dying cells, and to exclude them from further analysis. Included in AutoFoci are basic statistical analysis functions. For example, the focus numbers per cell are graphically compared with the theoretical Poisson distribution. In addition, they are compared by Kullback-Leibler (KL) divergence and the sum of the squared residuals respectively. The handling of the software is intuitive and well documented.
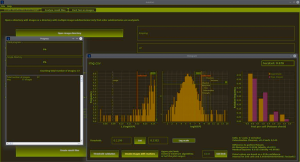
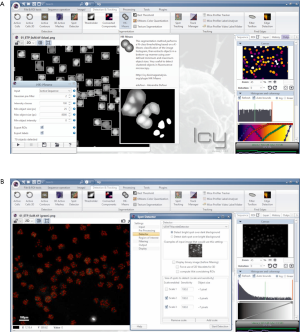
Icy—an open source bioimage informatics platform for reproducible research
Icy is an open platform with a comprehensive graphical interface for extended reproducible research in bioimage informatics. It is a free and user-oriented solution that non-expert users can run to change workflows according to their needs through graphical programming. This is supported by a community website (http://icy.bioimageanalysis.org/) that provides a centralized and open-access public repository to contribute and share plug-ins and workflows, and facilitates the development and usage of a variety of image processing algorithms (91,102).
Icy offers several plug-ins for different biological issues, protocols and scripts for adaptation of desired program settings. Tutorials and a suitable documentation at the Icy homepage facilitate the usage of plug-ins, scripts or protocols. Furthermore, Icy can be connected to Matlab and provides a native ImageJ integration. For the recognition and quantification of objects like nuclei or foci, segmentation methods and spot detection are available. Manual counting can also be performed via a plug-in. Thus, it is possible with the integrated plug-in HK Means to identify the nuclei by segmentation using user-defined minimum and maximum sizes of detected objects. This is also very useful to ascertain clustered objects in fluorescence microscopy. The output of the nuclei position makes it possible to determine the required ROI for the following foci detection. For foci detection, the Spot Detector tool can be used to recognize foci dependent on their area (Figure 5).
CellProfiler—user-friendly next-generation bioimage processing
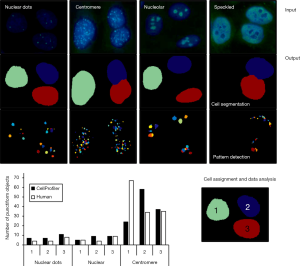
CellProfiler is an open source software to quantitatively measure phenotypes. The software has a flexible, modular design with a GUI. Sophisticated image analysis pipelines can be created for processing of large cellular image data to address complex biological questions even for non-programmers using CellProfiler and CellProfiler Analyst (Figure 6) (89,97). The release of CellProfiler 3.0 included improved support for both whole-volume and plane-wise analysis of three-dimensional image z-stacks (92). Custom pipelines can be created from modules that encapsulate e.g., image processing algorithms or object detection analysis (89,102). Functioning analysis pipelines are available on the developers’ homepage (http://cellprofiler.org/) as building blocks for customized pipelines.
As of completion of this review, there was no specialized module for foci detection. The Speckle Counting pipeline can be used instead. This advanced pipeline enables to identify smaller objects (foci) within larger objects (nuclei). Relationships between the two can be established as well as per-object aggregate measurements. Thus, nuclei and their contained foci can be characterized simultaneously (Figure 7).
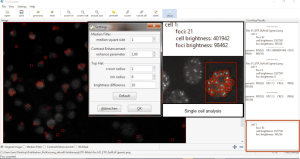
FociCounter—quantitative and qualitative analysis of γH2AX foci
FociCounter is a simple standalone program with a user-friendly GUI to obtain an overview about foci numbers. FociCounter is suitable for the foci determination within a cell, because the foci detection depends on brightness differences between the foci and the background (e.g., brightness of cytoplasm or nuclei) within a selected ROI (Figure 8). FociCounter as a semi-automatic program requires manual input for nuclei detection. Semi-automated computational tools with parameterized algorithms are most frequently presented in the literature (63). FociCounter is fairly minimalistic since it lacks features like automatic nuclei detection and the analysis of overlapping nuclei (44).
Commonly used open source programming languages with bioimage informatics capabilities
Programming languages, such as Python or R, are tools to build customized bioimage analysis pipelines. Both are widely adapted for scientific programming and data analysis. Their broad applicability and mathematical abilities makes them useful for big data mining, statistics and visualization (98). GUIs and integrated development environments (IDE) for image processing tasks have also been developed. They cover different standard functions such as thresholding, segmentation and different transformations. Bio7 (103) is a GUI and IDE which wraps the image analysis functionality of ImageJ and R. Single R packages for image analysis include EBImage (104) raster (105) and ripa (106), imager (107) and dcemir (108). Similarly, image processing can be performed in Python via scikit-image (109), OpenCV (110) or the Python Imaging Library (PIL) (111).
An example for the application Python for custom processing and analysis of immunofluorescence images is shown in Figure 9 (112). Thereby, nuclei were detected and the average intensity of both, the red and green signal inside the nucleus, was determined.
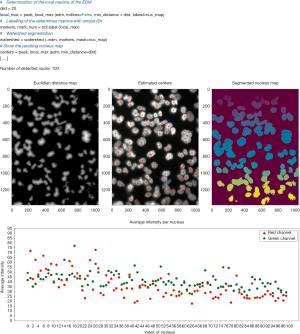
Challenges of bioimage informatics for the analysis of DNA damage
There are several challenges to humans that can be handled by bioimage informatics:
- The detection of punctiform objects in the nuclei as DSBs is demanding. The clustering of genes to nuclear bodies like PML bodies, speckles or Cajal bodies also cause foci formation (113). Furthermore, it is reported that PML bodies can also co-localize with γH2AX in DDR (114).
- Manual counting of cells that contain dozens of foci per cell (which may even overlap) and concomitant assessment of cellular structures is unsustainable as it is prone to inaccuracies and biases by the investigator and also yields low throughput (21,61). It also depends on the perception of the operator which may vary between different operators (71). This approach only allows a statement about the quantity of γH2AX foci for the specific image scenario rather than a generalized statement, because every image has to be individually adjusted for variations in focus and illumination (22,41,115). This was the case for all reviewed software.
- Dense and overlapping foci are quantifiable as long as the fluorescence signal is unsaturated and dedicated algorithms are used as described in (71).
- γH2AX/53BP1 foci and foci streaks may differ in their morphology, which requires more sophisticated algorithms (116).
- A crucial point is the robustness of chosen parameters for identification of nuclei and foci, which is strongly affected in the process of image acquisition and analysis (76).
- The variation in intensity of staining and the visual fusion of nuclear structures make it difficult to obtain an exact statement about the severity of the DNA damage by γH2AX foci analysis (76).
- Overlay methods help to generate visual estimates of co-localization events in two-dimensional images, but cannot reflect the three-dimensional nature of the biological sample (45).
- Confounders influencing the cell and foci detection, such as exposure time, the focus plane, cell debris and background signals.
- The direct estimation of the object density in an image without performing segmentation or object detection is difficult. This was realized elegantly in the software program ilastik, which represents an approach for counting blob-like overlapping objects with similar appearance (e.g., size, intensity, texture) (94). Overlapping foci can also be analyzed in more detail by 3D imaging. The vast amount of data generated by medical informatics and other research disciplines is an increasing challenge nowadays (98). The analysis of this so-called Big Data offers new research opportunities and holds the promise of improved quality of healthcare. The increasing volume of image data requires grid architectures to store, retrieve and process images of various formats among various entities (e.g., research units, hospitals) (108,117-119). Herein, efficient and interactive systems for the visualization of large data sets are needed. ImageJS is a browser-based computational ecosystem for an open, collaborative, computational image analysis (e.g., segmentation, feature extraction and filtering) that are both self-sustained and user driven (120). BigDataViewer was developed as a Fiji plug-in for interactive visualization of large 3D image data sets (121).
Digital image analysis and dedicated software packages allow the automated recognition of healthy and pathological cell structures and disease-related patterns (e.g., autoimmune diseases) and might create the basis for introduction of DDR markers into clinical routine. One way to improve the consistency of analysis in different images may be the training of supervised machine learning algorithms with user-labeled images (63,84). None of the software included such functionality.
Processing of γH2AX foci counts and recommendations
The following section aims to give recommendation for working with image data of DDR.
Errors (e.g., uneven illumination, blurriness, noise) are introduced during the measurement by microscope hardware (e.g., CCD camera), the specimen and data processing (46,122). Moreover, the intensity value of a pixel is not necessarily related to the number of fluorophores (mediated by a detector molecule like fluorescent antibodies) present since self-quenching, focusing, registration accuracy, bleed-through, auto-fluorescence, photo-bleaching and blinking effects disturb the signal (46,123-125). Additionally, the hardware-limited image resolution and the pixel aspect ratio do not allow for a one pixel per fluorophore ratio. It is better to assume that a single pixel represents the signal of multiple fluorophores.
- Differences such as staining intensity, small changes of sizes (e.g., nucleus size) or features like the texture are hard to detect by the human eye (89). Consequently, it is important to build models which describe rare repair foci events and to understand the corresponding DDR mechanisms. The latter requires large numbers of cells to ascertain such rare events, which may represent a subpopulation within a sample (52,89).
- Differences in foci morphology can be addressed by different methods. One example is High-content image-based cytometry (HCIC), which is a method to accurately quantify cellular characteristics (52,126). HCIC was used by Jezkova et al. to analyze γH2AX/53BP1 foci and foci streaks induced by high-LET boron and neon ions. They found that foci differed both in sizes and shapes depending on the LET radiation type (116). Another example is the usage of digital cameras in combination with confocal microscopes with deconvolution software enabled 3D image generation to get valuable information about foci.
- The number of cells analyzed is important for a well-founded statistical analysis. Although low cell counts reduce the amount of work required for manual counting, the cell count determines the accuracy of the assay. None of the reviewed software packages proposed criteria for this and thus, the definition of an adequate number of cells to analyze is the user’s responsibility. Therefore, the foci number may vary and could range from 50 to 1,000 (48). An analysis of 100 representative cells per condition could be suffice to draw statistically valid conclusions. We are not aware of a study that addressed this question. Intra- and inter-laboratory studies showed that huge differences in foci yields were obtained for the same samples. This was in part explained by variations in foci loss during shipment of blood samples or by variations in immunofluorescence staining (24).
- Over-dispersion, which means an excess of foci negative cells, is a possible scenario. In case of low cell numbers, over-dispersion cannot be observed.
- Since the foci number depends on variable properties like the foci fluorescence intensity (number of accumulated γH2AX), foci diameter, foci position, overlapping nuclei, background fluorescence and the occurrence of artifacts, automated focusing appears to be a prerequisite for robust and reproducible analysis pipelines.
- The lateral and z-axis resolution should be configured in a manner which suffices for analysis. Particularly the evaluation of an adequate z-axis resolution and a high enough lateral resolution for foci detection is demanding. It is important that an algorithm is capable of separating foci that are in close proximity or in contact with each other. Significant overlap of foci is likely with increasing dose of the DSB inducing agent by overall increasing the amount and thus density of foci (21).
- The standardization of γH2AX foci counting with meaningful biological controls is an essential requirement for the introduction into routine medicine (24,127).
- Bystander effects, technical influences during the measurement (e.g., noise, blurriness, inhomogeneous illumination) and biological variability require the assessment of hundreds of cells to exclude false-positive results. Several technical approaches have been introduced to perform an automatized image capturing and image quality management (69,128,129).
- γH2AX foci can be induced in adjacent cells, called bystander cells, by DNA damage through cell communication (130). Recognition and detection of foci are also used to investigate cellular architecture by fluorescence in-situ hybridization (FISH). Such foci are referred as FISH foci.
- Besides the foci data, it is important to analyze additional parameters. It was for example reported that pericentrin, an integral component of the centrosome, can be used to distinguish both the number of γ-H2AX foci per cell, and the cell cycle phase in a single assay (34).
- Different approaches were proposed to address the problem of overlapping foci (21). To segment overlapping foci, it was proposed to treat them as entities with a sufficient distance to clearly separate them (46). Several methods have been proposed to address uneven illumination, which is a serious challenge during image analysis. The same holds true for uneven illumination in light sheet microscopy (LSM) images (131,132). Uddin et al. presented a modified radiative transfer theory approach as alternative to deconvolution- or model-based restoration. They showed that the image restoration eliminated the contrast degradation problem of LSM (133). From the reviewed software only CellProfiler offered several methods to compensate uneven illumination (89).
Analysis of spatiotemporal foci relations and rejoining kinetics
Most studies appear to focus on the analysis of foci counts and co-localizations of DDR biomarkers. Direct visualization of the three-dimensional distribution of foci in the nucleus can additionally provide insight in the spatial organization of genome associated proteins and their association with cellular processes (e.g., gene expression) (34,134). The analysis of spatiotemporal foci relations and rejoining kinetics is important to understand DDR kinetics. For this task, different assays and software were developed accordingly (38,39,115,128,135,136). Studies in mammalian cells have suggested that both the yield and the spatial distribution of DSBs are influenced by various factors like the radiation quality (130).
An in the year 2005 by Desai et al. performed qualitative analysis provided insights into DNA damage processing kinetics for high charge and energy ions (134). This approach was used 2012 by Mok and Henderson to show that γH2AX-MDC1-53BP1 and RNF8-RNF168-BRCA1-A complexes were spatially independent (79). This provides important information about the spatial distribution of DSBs which needs to be considered in precision medicine.
Software tools, which are capable to analyze such data, might be also convenient for use in other fields of research. Genome editing by either zinc-finger nucleases or the CRISPR/Cas9 system also results in DSBs (137,138). Here, knowledge about spatial relationship of DSBs and known associated proteins and cell structures is certainly useful (Figure 10).
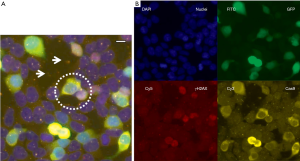
Example: PA28γ as a biomarker for pathophysiological conditions—difficulties and opportunities
The following paragraph describes challenges that a one might face during the analysis of image data. First, the biomedical hypothesis is explained and then linked to selected steps of analysis by bioimage informatics.
Proteasome activator PA28γ has initially been discovered as an autoantigen in systemic lupus erythematosus (SLE) (139,140) and has recently attracted attention as a putative biomarker in cancer (141,142). As for other biomarkers, the dynamics of expression, localization, either sub-cellularly or extra-cellularly, as well as the elicitation of autoantibodies derived against this protein, promotes the idea of using it as indicator of pathophysiological conditions or control parameter for monitoring of therapies. A proteomic approach revealed increased PA28γ expression in colorectal cancer (141). Several more recent studies indicated elevated PA28γ protein levels as a hallmark of epithelial or mesenchymal cancers (141,143-146). Since PA28γ localizes to sites of DSB and DDR, imaging approaches correlating PA28γ levels with therapeutic approaches inducing genotoxic damage might be of particular interest in the future.
The putative value of PA28γ as a bio-or-pathomarker was evaluated in sera of patients suffering from auto-immune diseases or cancer. They were examined for elevated levels of the PA28γ antigen and auto-antibodies. The parallel assessment of PA28γ antigen with a sandwich ELISA and autoantibodies with a microbead assay allowed assessing the putative competition of the ELISA antibody (detection limit 3 ng/mL) with autoantibodies, correlating with the antigen levels in RA sera. Intriguingly, high levels and diversity of autoantibodies may hamper quantification of amount of the corresponding PA28γ antigen. Such technical hindrances can be avoided in immunofluorescence imaging approaches, as discussed later. The design of the ELISA study suggests that PA28γ sera levels are elevated in cancer and autoimmune diseases. With regard to this antigen and the analytical approach, the differentiation between several forms of RA and cancer was limited, if not combined with other markers in multiparametric approaches. However, applied as a marker for surveillance of RA disease activity and therapy of rheumatoid arthritis, monitoring of PA28γ allowed correlation with therapeutic impact (142).
Monitoring of PA28γ protein or antibody sera levels may be of use in certain conditions. The diagnostic potential of PA28γ needs further studies. Since the molecular biology of PA28γ appears to be more sophisticated, future efforts should integrate such knowledge in imaging analysis on the subcellular distribution of post-translationally modified variants. PA28γ is described to be mainly localized in the nucleus. It can be translocated into the cytosol after SUMOylation at multiple sites (147). Interestingly, PA28γ is involved in multiple cellular processes related to cancer development such as apoptosis (148), cell cycle regulation, or the DNA damage response DDR (149,150). The functional state of PA28γ appears to be regulated post-translationally. Checkpoint kinase Chk2 phosphorylates PA28γ at serine 247, if switched on due to DNA damage PA28γ as an ATM phosphorylation target is recruited to DNA damage sites to enable the rapid accumulation of proteasomes at these sites. PA28γ-depended recruitment of 20S proteasomes has been shown by live cell imaging (151,152). PA28γ depletion enhanced the focal concentration of some proteins of the DNA replication apparatus at DNA damage sites. Contrarily, early focal dynamics represented by initial appearance of γH2AX seems to be independent of the presence of PA28γ, either indicating that PA28γ is involved in DDR protein dynamics at later states or that γH2AX levels are not affected by PA28γ-proteasome complexes.
Thus, PA28γ appears to be an additional biomarker, possibly not only for demonstrating sites of DNA damage, but for monitoring protein dynamics of repair at DNA damage sites. Several authors suggest that PA28γ plays a role in the coordination of the DNA double-strand repair and in chromosomal stability (151-153), but multiparametric analysis of protein dynamics that DNA damage and repair sites has so far not been considered for diagnostic purposes. Therefore, we established cellular models with CRISPR/Cas9-modulated PA28γ protein levels to investigate the role of PA28γ in DDR (154).
In Figure 11, a B8 fibroblast cell line overexpressing PA28γ (B8γ) has been compared with the B8 vector control (B8vc) regarding the subcellular distribution of PA28γ in relation to γH2AX after UV-C induction of DNA damage and apoptosis (148). In this in vitro we observed that PA28γ is preferentially located in the cytoplasm of B8vc fibroblasts, whereas overexpressing cells reveal a higher nuclear concentration, as well as a shift towards higher molecular weight complexes. These observations are confirmed by nuclear PA28γ translocation demonstrated by microscopic image analysis shown in Figure 11. Future studies on the correlations of PA28γ levels with markers of DSB foci and protein dynamics in the DDR protein signaling network should reveal, if PA28γ levels in cancer tissue affect the sensitivity of tissue towards genotoxic therapeutic intervention, abundancy of DSBs, kinetics of DSB appearance or repair.
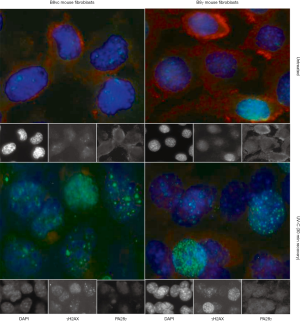
Conclusions
Semi-automatic detection and quantification of DSB markers uses automatic acquisition of multispectral images and analysis by bioimage informatics. Reproducible and reliable foci quantification is essential for data interpretation and for biological inferences. Therefore, different software packages for analysis of multispectral images were surveyed.
For users lacking profound knowledge of bioimage informatics, the availability of graphical user interfaces, which offer straightforward workflows, is an important prerequisite for entering digital image analysis. From our experience, all software packages fulfilled these perquisites and were eligible for reproducible quantification. When we handed out the software to inexperienced individuals [biotechnologists at bachelor level (N=10)], we received the (subjective, unrepresentative) feedback that the software was intuitive. CellProfiler and Icy were reported to provide the best user experience after a training phase. Lapytsko et al. found in their particular experimental setting (“time series of γ-irradiated cells for up to 10 Gy”) that CellProfiler and ImageJ have short comings (61). They reported a “poor performance on distributed foci”, “poor performance on images with low signal/noise ratio”, “poor performance on images with varying background” and that the both tools are “complicated to use”. We think that users need to evaluate software in small pilot studies before starting larger studies.
The aforementioned softwares were linked with numerous video and text tutorials, which eases their use. In general, the documentation was comprehensive for all software’s reviewed. More advanced users will appreciate the integrated development environments of ImageJ, CellProfiler and Icy, which empower them to extend the analysis and report generation routines by programming in Java.
We noticed that on several occasions’ knowledge of image processing termini was beneficial for ImageJ and CellProfiler to adjust the settings for successful object detection. Therefore, we would like to point the readers of this review to the excellent books by Jähne and Burger, respectively (65,155).
Most software can recognize and measure patterns and phenotypes by dedicated plug-ins [e.g., Icy (91)], macros [e.g., ImageJ (85)] or pipelines [e.g., CellProfiler (89)]. Semi-automated computational tools with parameterized algorithms are most frequently presented in the literature (63). CellProfiler uses the concept of pipelines, which are usable to analyze cell patterns besides the foci. CellProfiler version 2.0 and later offers user documentation and a pipeline called “Speckle Counting” that is suitable for foci counting. The visualization of the location, frequency and in particular the co-localization with other biomolecules (e.g., via miRNA, FISH) may be a meaningful addition on the phenotypic level. In contrast, FociCounter lacks this and other features like automatic nuclei detection and segmentation of overlapping nuclei. In consequence, the scope of FociCounter is limited to the enumeration of foci and nuclei. This limits its applicability in precision medicine.
The classification of γH2AX foci is achieved through intensity and size analysis of the fluorescent spots. The open bioimage informatics platforms Icy and CellProfiler were most appropriate in terms of usability and analysis robustness. Except for FociCounter, it was possible to simultaneously analyze other DSB associated biomarkers (e.g., 53BP1, nucleus size).
Conventional approaches use static instructions to obtain information from an image. For example, most ImageJ plug-ins use intensity thresholding and a given minimal size to distinguish foci from the background. As with all automatic analysis, there is a trade-off between exact foci quantification and throughput. When unsupervised, the algorithms identified not all individual nuclei and foci and may recognize cell debris as foci in complex images. The analysis quality was largely depended on the user-defined thresholds. Therefore, when used in scientific studies, it is important to state this information precisely for reproducible research. An alternative approach is machine learning, which could be used to build a statistical model from the image data. The generated models could further be used to make predictions and decisions (156,157). Machine learning for example was used for the analysis of large datasets and multivariate phenotypic profiling for example to determine differing DDR related patterns (126).
After digital image analysis, statistical processing, data storage and report generation follows. γH2AX foci are discrete punctiform objects that can be quantified by counting. Only AutoFoci offered basic statistical analysis modules for count data (e.g., Poisson statistics). Most of them gave information about the average number of foci per cell. This is problematic since many datasets contain cells with no foci at all.
Foci signal intensities, their spatial distribution (2D, 3D) as well as time-dependent signals can also be quantified by certain software (78,79). It is important that researchers revise the capabilities of the software intended to be used.
High resolved images are problematic regarding deconvolution techniques and complexity of the image. In the case of punctiform objects like foci, this complexity can cause artificial clustering and over-counting. Reasons for the latter are over-labeling and physical cross-linked proteins via antibodies (158). Therefore, attention needs to be paid to ostensible co-localization of foci (e.g., γH2AX and 53BP1).
Regarding the complexity of DNA damage repair mechanism, a meaningful definition of what a focus actually means in the context of DNA damage needs to be found, especially since the functional relevance of foci as well as the chain of protein phosphorylation and ubiquitination at chromatin level is not fully understood.
Understandably, in the context of a review not all available software can be investigated. Nevertheless, we hope to provide a good overview of analysis software in context of DNA repair foci characterization.
Acknowledgments
Funding: This work has in part been funded by the “Gesundheitscampus Brandenburg - Konsequenzen der altersassoziierten Zell - und Organfunktionen” initiative of the Brandenburgian Ministry of Science, Research and Culture (MWFK) and in part by the InnoProfile-Transfer 03 IPT611X project of the Federal Ministry of Education and Research (BMBF).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “DNA Damage Assessment for Precision Medicine”. The article has undergone external peer review.
Conflicts of Interest: The series “DNA Damage Assessment for Precision Medicine” was commissioned by the editorial office without any funding or sponsorship. Dirk Roggenbuck served as an unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Laboratory and Precision Medicine from August 2017 to July 2019. Dirk Roggenbuck is a shareholder and employee of GA Generic Assays GmbH and Medipan GmbH. Other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003;3:155-68. [Crossref] [PubMed]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001;27:247-54. [Crossref] [PubMed]
- Krokan HE, Bjørås M. Base Excision Repair. Cold Spring Harb Perspect Biol 2013;5:a012583 [Crossref] [PubMed]
- Li G-M. Mechanisms and functions of DNA mismatch repair. Cell Res 2008;18:85-98. [Crossref] [PubMed]
- Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183-235. [Crossref] [PubMed]
- Scherthan H, Hieber L, Braselmann H, et al. Accumulation of DSBs in γ-H2AX domains fuel chromosomal aberrations. Biochem Biophys Res Commun 2008;371:694-7. [Crossref] [PubMed]
- Bonner WM, Redon CE, Dickey JS, et al. γH2AX and cancer. Nat Rev Cancer 2008;8:957-67. [Crossref] [PubMed]
- Guillerman RP. From ‘Image Gently’ to image intelligently: a personalized perspective on diagnostic radiation risk. Pediatr Radiol. 2014;44:444-9. [Crossref] [PubMed]
- Rödiger S, Liefold M, Ruhe M, et al. Quantification of DNA double-strand breaks in peripheral blood mononuclear cells from healthy donors exposed to bendamustine by an automated γH2AX assay—an exploratory study. J Lab Precis Med 2018;3:47. [Crossref]
- Ruhe M, Rabe D, Jurischka C, et al. Molecular biomarkers of DNA damage in diffuse large-cell lymphoma—a review. J Lab Precis Med 2019;4:5. [Crossref]
- Rogakou EP, Pilch DR, Orr AH, et al. DNA Double-stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J Biol Chem 1998;273:5858-68. [Crossref] [PubMed]
- Rogakou EP, Boon C, Redon C, et al. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo. J Cell Biol 1999;146:905-16. [Crossref] [PubMed]
- Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A 2003;100:5057-62. [Crossref] [PubMed]
- Shamir L, Delaney JD, Orlov N, et al. Pattern Recognition Software and Techniques for Biological Image Analysis. PLoS Comput Biol 2010;6:e1000974 [Crossref] [PubMed]
- Ambers A, Turnbough M, Benjamin R, et al. Assessment of the role of DNA repair in damaged forensic samples. Int J Legal Med 2014;128:913-21. [Crossref] [PubMed]
- Shukla RK. Forensic application of comet assay: an emerging technique. Forensic Sci Res 2017;2:180-4. [Crossref] [PubMed]
- Diegoli TM, Farr M, Cromartie C, et al. An optimized protocol for forensic application of the PreCRTM Repair Mix to multiplex STR amplification of UV-damaged DNA. Forensic Science International: Genetics 2012;6:498-503. [Crossref] [PubMed]
- Hall A, Ballantyne J. Characterization of UVC-induced DNA damage in bloodstains: forensic implications. Anal Bioanal Chem 2004;380:72-83. [Crossref] [PubMed]
- Vasireddy RS, Karagiannis TC, El-Osta A. γ-radiation-induced γH2AX formation occurs preferentially in actively transcribing euchromatic loci. Cell Mol Life Sci 2010;67:291-4. [Crossref] [PubMed]
- Vieira MLC, Santini L, Diniz AL, et al. Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 2016;39:312-28. [Crossref] [PubMed]
- Böcker W, Iliakis G. Computational Methods for analysis of foci: validation for radiation-induced gamma-H2AX foci in human cells. Radiat Res 2006;165:113-24. [Crossref] [PubMed]
- Jucha A, Wegierek-Ciuk A, Koza Z, et al. FociCounter: A freely available PC programme for quantitative and qualitative analysis of gamma-H2AX foci. Mutat Res 2010;696:16-20. [Crossref] [PubMed]
- Anderson D, Andrais B, Mirzayans R, et al. Comparison of two methods for measuring γ-H2AX nuclear fluorescence as a marker of DNA damage in cultured human cells: applications for microbeam radiation therapy. J Instrum 2013;8:C06008 [Crossref]
- Rothkamm K, Horn S, Scherthan H, et al. Laboratory Intercomparison on the γ-H2AX Foci Assay. Radiat Res 2013;180:149-55. [Crossref] [PubMed]
- Reddig A, Rübe CE, Rödiger S, et al. DNA damage assessment and potential applications in laboratory diagnostics and precision medicine. J Lab Precis Med 2018;3:31. [Crossref]
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687-96. [Crossref] [PubMed]
- Pilch DR, Sedelnikova OA, Redon C, et al. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol 2003;81:123-9. [Crossref] [PubMed]
- Luczak MW, Zhitkovich A. Monoubiquitinated γ-H2AX: Abundant product and specific biomarker for non-apoptotic DNA double-strand breaks. Toxicol Appl Pharmacol 2018;355:238-46. [Crossref] [PubMed]
- Eberlein U, Peper M, Fernández M, et al. Calibration of the γ-H2AX DNA Double Strand Break Focus Assay for Internal Radiation Exposure of Blood Lymphocytes. PLoS One 2015;10:e0123174 [Crossref] [PubMed]
- Marková E, Schultz N, Belyaev IY. Kinetics and dose-response of residual 53BP1/gamma-H2AX foci: co-localization, relationship with DSB repair and clonogenic survival. Int J Radiat Biol 2007;83:319-29. [Crossref] [PubMed]
- Rappold I, Iwabuchi K, Date T, et al. Tumor Suppressor P53 Binding Protein 1 (53bp1) Is Involved in DNA Damage-Signaling Pathways. J Cell Biol 2001;153:613-20. [Crossref] [PubMed]
- Kang K, Lee SB, Yoo J-H, et al. Flow cytometric fluorescence pulse width analysis of etoposide-induced nuclear enlargement in HCT116 cells. Biotechnol Lett 2010;32:1045-52. [Crossref] [PubMed]
- Löbrich M, Shibata A, Beucher A, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 2010;9:662-9. [Crossref] [PubMed]
- Hernández L, Terradas M, Martín M, et al. Highly Sensitive Automated Method for DNA Damage Assessment: Gamma-H2AX Foci Counting and Cell Cycle Sorting. Int J Mol Sci 2013;14:15810-26. [Crossref] [PubMed]
- Bouley J, Saad L, Grall R, et al. A new phosphorylated form of Ku70 identified in resistant leukemic cells confers fast but unfaithful DNA repair in cancer cell lines. Oncotarget 2015;6:27980-8000. [Crossref] [PubMed]
- Bekker-Jensen S, Danielsen JR, Fugger K, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 2010;12:80-6. Corrected in Nat Cell Biol 2010;12:412. [Crossref] [PubMed]
- Paull TT, Rogakou EP, Yamazaki V, et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000;10:886-95. [Crossref] [PubMed]
- Padfield D, Rittscher J, Thomas N, et al. Spatio-temporal cell cycle phase analysis using level sets and fast marching methods. Med Image Anal 2009;13:143-55. [Crossref] [PubMed]
- Turner HC, Sharma P, Perrier JR, et al. The RABiT: High Throughput Technology for Assessing Global DSB Repair. Radiat Environ Biophys 2014;53:265-72. [Crossref] [PubMed]
- Barni MV, Carlini MJ, Cafferata EG, et al. Carnosic acid inhibits the proliferation and migration capacity of human colorectal cancer cells. Oncol Rep 2012;27:1041-8. [Crossref] [PubMed]
- Bhogal N, Jalali F, Bristow RG. Microscopic imaging of DNA repair foci in irradiated normal tissues. Int J Radiat Biol 2009;85:732-46. [Crossref] [PubMed]
- Willitzki A, Hiemann R, Peters V, et al. New Platform Technology for Comprehensive Serological Diagnostics of Autoimmune Diseases, New Platform Technology for Comprehensive Serological Diagnostics of Autoimmune Diseases. Clin Dev Immunol 2012;2012:e284740 [Crossref] [PubMed]
- Şener AG, Afşar İ. Frequency of dense fine speckled pattern in immunofluorescence screening test. Eur J Rheumatol 2015;2:103-5. [Crossref] [PubMed]
- González JE, Radl A, Romero I, et al. Automatic Detection of Mitosis and Nuclei From Cytogenetic Images by CellProfiler Software for Mitotic Index Estimation. Radiat Prot Dosimetry 2016;172:218-22. [Crossref] [PubMed]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 2006;224:213-32. [Crossref] [PubMed]
- Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol 2009;185:1135-48. [Crossref] [PubMed]
- Reddig A, Roggenbuck D, Reinhold D. Comparison of different immunoassays for γH2AX quantification. J Lab Precis Med 2018;3:80. [Crossref]
- Garty G, Bigelow AW, Repin M, et al. An automated imaging system for radiation biodosimetry. Microsc Res Tech 2015;78:587-98. [Crossref] [PubMed]
- Moreno-Villanueva M, Pfeiffer R, Sindlinger T, et al. A modified and automated version of the “Fluorimetric Detection of Alkaline DNA Unwinding” method to quantify formation and repair of DNA strand breaks. BMC Biotechnol 2009;9:39. [Crossref] [PubMed]
- Baumstark-Khan C, Hentschel U, Nikandrova Y, et al. Fluorometric Analysis of DNA Unwinding (FADU) as a Method for Detecting Repair-induced DNA Strand Breaks in UV-irradiated Mammalian Cells¶. Photochem Photobiol 2000;72:477-84. [Crossref] [PubMed]
- Khan K, Tewari S, Awasthi NP, et al. Flow cytometric detection of gamma-H2AX to evaluate DNA damage by low dose diagnostic irradiation. Med Hypotheses 2018;115:22-8. [Crossref] [PubMed]
- Wadduwage DN, Parrish M, Choi H, et al. Subnuclear foci quantification using high-throughput 3D image cytometry. Available online: http://dx.doi.org/
10.1117/12.2185005 - Lim D, Chu KK, Mertz J. Wide-field fluorescence sectioning with hybrid speckle and uniform-illumination microscopy. Opt Lett 2008;33:1819. [Crossref] [PubMed]
- Choi H, Wadduwage DN, Tu TY, et al. Three-Dimensional Image Cytometer Based on Widefield Structured Light Microscopy and High-Speed Remote Depth Scanning. Cytometry A 2015;87:49-60. [Crossref] [PubMed]
- Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006;1:23-9. [Crossref] [PubMed]
- Bajpayee M, Kumar A, Dhawan A. The Comet Assay: Assessment of In Vitro and In Vivo DNA Damage. In: Dhawan A, Bajpayee M, editors. Genotoxicity Assessment. Methods in Molecular Biology (Methods and Protocols), vol 1044. Humana Press, Totowa, NJ.
- Apostolou P, Toloudi M, Kourtidou E, et al. Use of the comet assay technique for quick and reliable prediction of in vitro response to chemotherapeutics in breast and colon cancer. J Biol Res-Thessalon 2014;21:14. [Crossref] [PubMed]
- Watson C, Ge J, Cohen J, et al. High-Throughput Screening Platform for Engineered Nanoparticle-Mediated Genotoxicity Using CometChip Technology. ACS Nano 2014;8:2118-33. [Crossref] [PubMed]
- Lovell DP, Omori T. Statistical issues in the use of the comet assay. Mutagenesis 2008;23:171-82. [Crossref] [PubMed]
- Helma C, Uhl M. A public domain image-analysis program for the single-cell gel-electrophoresis (comet) assay. Mutat Res 2000;466:9-15. [Crossref] [PubMed]
- Lapytsko A, Kollarovic G, Ivanova L, et al. FoCo: a simple and robust quantification algorithm of nuclear foci. BMC Bioinformatics 2015;16:392. [Crossref] [PubMed]
- Caicedo JC, Roth J, Goodman A, et al. Evaluation of Deep Learning Strategies for Nucleus Segmentation in Fluorescence Images. bioRxiv 2018;335216: [Crossref]
- Herbert AD, Carr AM, Hoffmann E. FindFoci: A Focus Detection Algorithm with Automated Parameter Training That Closely Matches Human Assignments, Reduces Human Inconsistencies and Increases Speed of Analysis. Lichten M, editor. PLoS One 2014;9:e114749.
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans Syst Man Cybern 1979;9:62-6. [Crossref]
- Jähne B. Digital Image Processing. 5th ed. Berlin Heidelberg: Springer-Verlag; 2002.
- Sklansky J. Image Segmentation and Feature Extraction. IEEE Trans Syst Man Cybern 1978;8:237-47. [Crossref]
- Loukas CG, Wilson GD, Vojnovic B, et al. An image analysis-based approach for automated counting of cancer cell nuclei in tissue sections. Cytometry A 2003;55:30-42. [Crossref] [PubMed]
- Zhang H, Fritts JE, Goldman SA. Image segmentation evaluation: A survey of unsupervised methods. Comput Vis Image Underst 2008;110:260-80. [Crossref]
- Willitzki A, Lorenz S, Hiemann R, et al. Fully automated analysis of chemically induced γH2AX foci in human peripheral blood mononuclear cells by indirect immunofluorescence. Cytometry A 2013;83:1017-26. [Crossref] [PubMed]
- Carpenter AE, Kamentsky L, Eliceiri KW. A call for bioimaging software usability. Nat Methods. 2012;9:666-70. [Crossref] [PubMed]
- Feng J, Lin J, Zhang P, et al. A novel automatic quantification method for high-content screening analysis of DNA double strand-break response. Sci Rep 2017;7:9581. [Crossref] [PubMed]
- Krause C, Ens K, Fechner K, et al. EUROPattern Suite technology for computer-aided immunofluorescence microscopy in autoantibody diagnostics. Lupus 2015;24:516-29. [Crossref] [PubMed]
- Schunck C, Johannes T, Varga D, et al. New developments in automated cytogenetic imaging: unattended scoring of dicentric chromosomes, micronuclei, single cell gel electrophoresis, and fluorescence signals. Cytogenet Genome Res 2004;104:383-9. [Crossref] [PubMed]
- Mahler M, Fritzler MJ. The Clinical Significance of the Dense Fine Speckled Immunofluorescence Pattern on HEp-2 Cells for the Diagnosis of Systemic Autoimmune Diseases. Clin Dev Immunol 2012;2012:494356 [Crossref] [PubMed]
- Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 2013;202:579-95. [Crossref] [PubMed]
- Ronneberger O, Baddeley D, Scheipl F, et al. Spatial quantitative analysis of fluorescently labeled nuclear structures: Problems, methods, pitfalls. Chromosome Res 2008;16:523. [Crossref] [PubMed]
- McVean A, Kent S, Bakanov A, et al. Development and validation of “AutoRIF”: software for the automated analysis of radiation-induced foci. Genome Integr 2012;3:1. [Crossref] [PubMed]
- Vasireddy RS, Tang MM, Mah LJ, et al. Evaluation of the Spatial Distribution of γH2AX following Ionizing Radiation. J Vis Exp 2010;2203. [PubMed]
- Mok MTS, Henderson BR. Three-dimensional imaging reveals the spatial separation of γH2AX-MDC1-53BP1 and RNF8-RNF168-BRCA1-A complexes at ionizing radiation-induced foci. Radiother Oncol 2012;103:415-20. [Crossref] [PubMed]
- Sowa M, Reddig A, Schierack P, et al. Phosphorylated histone 2AX foci determination in capillary blood mononuclear cells. J Lab Precis Med 2018;3:45. [Crossref]
- Lee JM, Gordon N, Trepel JB, et al. Development of a multiparameter flow cytometric assay as a potential biomarker for homologous recombination deficiency in women with high-grade serous ovarian cancer. J Transl Med 2015;13:239. [Crossref] [PubMed]
- Nakamura AJ, Redon CE, Bonner WM, et al. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging 2009;1:212-8. [Crossref] [PubMed]
- Eliceiri KW, Berthold MR, Goldberg IG, et al. Biological imaging software tools. Nat Methods 2012;9:697-710. Corrected in Nat Methods 2012;9(:1031. [Crossref] [PubMed]
- Sommer C, Gerlich DW. Machine learning in cell biology - teaching computers to recognize phenotypes. J Cell Sci 2013;126:5529-39. [Crossref] [PubMed]
- Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int 2004;11:36-42.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671-5. [Crossref] [PubMed]
- Oeck S, Malewicz NM, Hurst S, et al. The Focinator - a new open-source tool for high-throughput foci evaluation of DNA damage. Radiat Oncol 2015;10:163. [Crossref] [PubMed]
- Oeck S, Malewicz NM, Hurst S, et al. The Focinator v2-0 - Graphical Interface, Four Channels, Colocalization Analysis and Cell Phase Identification. Radiat Res 2017;188:114-20. [Crossref] [PubMed]
- Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7:R100. [Crossref] [PubMed]
- Deutzmann A, Ganz M, Schönenberger F, et al. The human oncoprotein and chromatin architectural factor DEK counteracts DNA replication stress. Oncogene 2015;34:4270-7. [Crossref] [PubMed]
- de Chaumont F. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 2012;9:690-6. [Crossref] [PubMed]
- McQuin C, Goodman A, Chernyshev V, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 2018;16:e2005970 [Crossref] [PubMed]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. BioTechniques 2007;42:71-5. [Crossref] [PubMed]
- Sommer C, Straehle C, Köthe U, et al. ilastik: Interactive Learning and Segmentation Toolkit. Proc 8th IEEE Int Symp Biomed Imaging Nano Macro. 2011 Jan 24;230-3. Available online: https://ieeexplore.ieee.org/document/5872394/citations#citations
- Lengert N, Mirsch J, Weimer RN, et al. AutoFoci, an automated high-throughput foci detection approach for analyzing low-dose DNA double-strand break repair. Sci Rep 2018;8:17282. [Crossref] [PubMed]
- Herbert A. IMAGEJ FINDFOCI Plugins [Internet]. University of Sussex; 2016 [cited 2018 Nov 23]. Available online: https://web.archive.org/web/20181123135032/http://www.sussex.ac.uk/gdsc/intranet/pdfs/FindFoci.pdf
- Jones TR, Kang IH, Wheeler DB, et al. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 2008;9:482. [Crossref] [PubMed]
- Chen M, Mao S, Liu Y. Big Data: A Survey. Mob Netw Appl 2014;19:171-209. [Crossref]
- Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676-82. [Crossref] [PubMed]
- Edelstein AD, Tsuchida MA, Amodaj N, et al. Advanced methods of microscope control using µManager software. J Biol Methods 2014;1:e10 [Crossref] [PubMed]
- Delibaltov DL, Gaur U, Kim J, et al. CellECT: cell evolution capturing tool. BMC Bioinformatics 2016;17:88. [Crossref] [PubMed]
- Wiesmann V, Franz D, Held C, et al. Review of free software tools for image analysis of fluorescence cell micrographs. J Microsc 2015;257:39-53. [Crossref] [PubMed]
- Austenfeld M, Beyschlag W. A Graphical User Interface for R in a Rich Client Platform for Ecological Modeling. J Stat Softw 2012;49:1-19. [Crossref]
- Pau G, Fuchs F, Sklyar O, et al. EBImage—an R package for image processing with applications to cellular phenotypes. Bioinformatics 2010;26:979-81. [Crossref] [PubMed]
- Hijmans RJ, Etten J van, Cheng J, et al. raster: Geographic Data Analysis and Modeling. Available online: https://cran.r-project.org/web/packages/raster/index.html
- Perciano T, Frery AC. ripa: R Image Processing and Analysis [Internet]. 2014. Available online: https://cran.r-project.org/web/packages/ripa/index.html
- Barthelmé, Simon. imager: an R package for image processing. Available online: https://dahtah.github.io/imager/
- R Package for the Analysis of DCE-MRI. SourceForge. Available online: http://sourceforge.net/projects/dcemri/
- van der Walt S, Schönberger JL, Nunez-Iglesias J, et al. scikit-image: image processing in Python. PeerJ 2014;2:e453 [Crossref] [PubMed]
- Bradski G. The OpenCV library. Available online: http://www.drdobbs.com/open-source/the-opencv-library/184404319.
-
Python Imaging Library (PIL) - SilMon. SilMon/DSB-Analysis-Notebook: v1.0. Available online: https://zenodo.org/record/2563812#.XGRP8VxKjIV
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295-306. [Crossref] [PubMed]
- Chang HR, Munkhjargal A, Kim MJ, et al. The functional roles of PML nuclear bodies in genome maintenance. Mutat Res 2018;809:99-107. [Crossref] [PubMed]
- Qvarnström OF, Simonsson M, Johansson KA, et al. DNA double strand break quantification in skin biopsies. Radiother Oncol 2004;72:311-7. [Crossref] [PubMed]
- Jezkova L, Zadneprianetc M, Kulikova E, et al. Particles with similar LET values generate DNA breaks of different complexity and reparability: a high-resolution microscopy analysis of γH2AX/53BP1 foci. Nanoscale 2018;10:1162-79. [Crossref] [PubMed]
- Herland M, Khoshgoftaar TM, Wald R. A review of data mining using big data in health informatics. J Big Data 2014;1:2. [Crossref]
- Zhang Z. The role of big-data in clinical studies in laboratory medicine. J Lab Precis Med 2017;2:34. [Crossref]
- Grace RK, Manimegalai R, Kumar SS. Medical Image Retrieval System in Grid Using Hadoop Framework. In: 2014 International Conference on Computational Science and Computational Intelligence (CSCI). 2014. p. 144-8.
- Almeida JS, Iriabho EE, Gorrepati VL, et al. ImageJS: Personalized, participated, pervasive, and reproducible image bioinformatics in the web browser. J Pathol Inform 2012;3:25. [Crossref] [PubMed]
- Pietzsch T, Saalfeld S, Preibisch S, et al. BigDataViewer: visualization and processing for large image data sets. Nat Methods 2015;12:481-3. [Crossref] [PubMed]
- Wählby C, Kamentsky L, Liu ZH, et al. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods 2012;9:714-6. [Crossref] [PubMed]
- Zwier JM, Van Rooij GJ, Hofstraat JW, et al. Image calibration in fluorescence microscopy. J Microsc 2004;216:15-24. [Crossref] [PubMed]
- Melan MA, Sluder G. Redistribution and differential extraction of soluble proteins in permeabilized cultured cells. Implications for immunofluorescence microscopy. J Cell Sci 1992;101:731-43. [PubMed]
- Gibson E, Fenster A, Ward AD. The impact of registration accuracy on imaging validation study design: A novel statistical power calculation. Med Image Anal 2013;17:805-15. [Crossref] [PubMed]
- De Vos WH, Van Neste L, Dieriks B, et al. High content image cytometry in the context of subnuclear organization. Cytometry A 2010;77:64-75. [PubMed]
- Rothkamm K, Barnard S, Moquet J, et al. DNA damage foci: Meaning and significance. Environ Mol Mutagen 2015;56:491-504. [Crossref] [PubMed]
- Barber PR, Locke RJ, Pierce GP, et al. Gamma-H2AX foci counting: image processing and control software for high-content screening. Available online: http://dx.doi.org/
10.1117/12.705217 - Rödiger S, Schierack P, Böhm A, et al. A Highly Versatile Microscope Imaging Technology Platform for the Multiplex Real-Time Detection of Biomolecules and Autoimmune Antibodies. In: Seitz H, Schumacher S, editors. Molecular Diagnostics. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012 p. 35-74.
- Prise KM, Folkard M, Kuosaite V, et al. What role for DNA damage and repair in the bystander response? Mutat Res 2006;597:1-4. [Crossref] [PubMed]
- Chen T, Yin W, Zhou XS, et al. Illumination normalization for face recognition and uneven background correction using total variation based image models. 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05). 20-25 June 2005. San Diego: IEEE; 2005.
- Sarder P, Nehorai A. Deconvolution methods for 3-D fluorescence microscopy images. IEEE Signal Process Mag 2006;23:32-45. [Crossref]
- Uddin MS, Lee HK, Preibisch S, et al. Restoration of uneven illumination in light sheet microscopy images. Microsc Microanal 2011;17:607-13. [Crossref] [PubMed]
- Desai N, Davis E, O’Neill P, et al. Immunofluorescence detection of clustered gamma-H2AX foci induced by HZE-particle radiation. Radiat Res 2005;164:518-22. [Crossref] [PubMed]
- Huggett J, O’Grady J, Bustin S. How to make Mathematics Biology’s next and better microscope. Biomol Detect Quantif 2014;1:A1-3. [Crossref] [PubMed]
- Hiraoka Y, Shimi T, Haraguchi T. Multispectral Imaging Fluorescence Microscopy for Living Cells. Cell Struct Funct 2002;27:367-74. [Crossref] [PubMed]
- Carroll D. Genome Engineering With Zinc-Finger Nucleases. Genetics. 2011;188:773-82. [Crossref] [PubMed]
- Lee S, Lee CY, Lee J, et al. Cut and paste the genome: Genome editing for research and therapy. J Cell Biotechnol 2015;1:95-106. [Crossref]
- Tojo T, Kaburaki J, Hayakawa M, et al. Precipitating antibody to a soluble nuclear antigen “Ki” with specificity for systemic lupus erythematosus. Ryumachi 1981;21:129-40. [PubMed]
- Nikaido T, Shimada K, Shibata M, et al. Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Clin Exp Immunol 1990;79:209-14. [Crossref] [PubMed]
- Roessler M, Rollinger W, Mantovani-Endl L, et al. Identification of PSME3 as a Novel Serum Tumor Marker for Colorectal Cancer by Combining Two-dimensional Polyacrylamide Gel Electrophoresis with a Strictly Mass Spectrometry-based Approach for Data Analysis. Mol Cell Proteomics 2006;5:2092-101. [Crossref] [PubMed]
- Gruner M, Moncsek A, Rödiger S, et al. Increased proteasome activator 28 gamma (PA28γ) levels are unspecific but correlate with disease activity in rheumatoid arthritis. BMC Musculoskelet Disord 2014;15:414. [Crossref] [PubMed]
- Kondo M, Moriishi K, Wada H, et al. Upregulation of nuclear PA28γ expression in cirrhosis and hepatocellular carcinoma. Exp Ther Med 2012;3:379-85. [Crossref] [PubMed]
- Kontos CK. Surrogate Prognostic Biomarkers in OSCC: The Paradigm of PA28γ Overexpression. EBioMedicine 2015;2:784-5. [Crossref] [PubMed]
- Wang X, Tu S, Tan J, et al. REG gamma: a potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med Oncol 2011;28:31-41. [Crossref] [PubMed]
- Chen D, Yang X, Huang L, et al. The Expression and Clinical Significance of PA28 γ in Colorectal Cancer. J Investig Med 2013;61:1192-6. [Crossref] [PubMed]
- Wu Y, Wang L, Zhou P, et al. Regulation of REGγ cellular distribution and function by SUMO modification. Cell Res 2011;21:807-16. [Crossref] [PubMed]
- Moncsek A, Gruner M, Meyer H, et al. Evidence for anti-apoptotic roles of proteasome activator 28γ via inhibiting caspase activity. Apoptosis. 2015;20:1211-28. [Crossref] [PubMed]
- Mao I, Liu J, Li X, et al. REGγ, a proteasome activator and beyond? Cell Mol Life Sci 2008;65:3971. [Crossref] [PubMed]
- Stohwasser R. Proteasome Activator 28γ: Impact on Survival Signaling and Apoptosis. In: Tutar Y, Tutar L, editors. Current Understanding of Apoptosis: Programmed Cell Death. 2018.
- Levy-Barda A, Lerenthal Y, Davis AJ, et al. Involvement of the nuclear proteasome activator PA28γ in the cellular response to DNA double-strand breaks. Cell Cycle. 2011;10:4300-10. [Crossref] [PubMed]
- Magni M, Ruscica V, Buscemi G, et al. Chk2 and REGγ-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res 2014;42:13150-60. [Crossref] [PubMed]
- Zannini L, Lecis D, Buscemi G, et al. REGγ proteasome activator is involved in the maintenance of chromosomal stability. Cell Cycle 2008;7:504-12. [Crossref] [PubMed]
- Rosanski M, Sauer L, Schröder C, et al. Knockout of proteolytic key regulators in malignant peripheral nerve sheath tumor cells by CRISPR/Cas9. J Cell Biotechnol 2019;4:5-13. [Crossref]
- Burger W, Burge MJ. Digital Image Processing: An Algorithmic Introduction Using Java. 2nd ed. London: Springer-Verlag; 2016.
- Kan A. Machine learning applications in cell image analysis. Immunol Cell Biol 2017;95:525-30. [Crossref] [PubMed]
- Wernick MN, Yang Y, Brankov JG, et al. Machine Learning in Medical Imaging. IEEE Signal Process Mag 2010;27:25-38. [Crossref] [PubMed]
- Durisic N, Cuervo LL, Lakadamyali M. Quantitative super-resolution microscopy: pitfalls and strategies for image analysis. Curr Opin Chem Biol 2014;20:22-8. [Crossref] [PubMed]
Cite this article as: Schneider J, Weiss R, Ruhe M, Jung T, Roggenbuck D, Stohwasser R, Schierack P, Rödiger S. Open source bioimage informatics tools for the analysis of DNA damage and associated biomarkers. J Lab Precis Med 2019;4:21.



