
Comparing the performance of two commercial assays for detection of SARS-CoV-2 in samples with low viral load
Dynamic change process of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gene targets can be monitored over time in individual patients with coronavirus disease 2019 (COVID-19) by real-time reverse transcription polymerase chain reaction (rRT-PCR) (1). In addition to confirming diagnosis, dual-target rRT-PCR assays are currently being used to generate cycle threshold (Ct) values in follow-up samples from COVID-19 patients (2). A study by Zóka et al. showed N gene may persist longer and we would like to share our experience with two dual-target SARS-CoV-2 molecular diagnostic tests with which samples with low viral load were tested.
For many diagnostic laboratories it is not unusual to use multiple validated assays for SARS-CoV-2 testing to cope with high testing demand since it will be too risky for a laboratory to “place all eggs in one basket”. More importantly, immediate availability of alternate assays will be crucial to serve as back-up methods to the primary assay to minimize interruption in service during machine break-down, temporary assay suspension due to contamination, or supply shortage of reagents or test kits. Another advantage of having an alternate assay is that different assays may target different parts of the viral genome, so that if there is an inconclusive result from one assay, the other assay could be used to resolve the uncertainty.
In our laboratory, we perform SARS-CoV-2 diagnostic tests using the cobas® SARS-CoV-2 test (Roche Molecular Systems, NJ, USA), a Food and Drug Administration (FDA) emergency use authorization (EUA) assay targeting the ORF1a and E gene targets, and another FDA EUA assay, the Xpert® Xpress SARS-Cov-2 assay (Cepheid, CA, USA). During the first 6 months of the pandemic patients were tested serially to determine when they can be safely discharged from isolation. We recently reviewed the Ct values from 24 consecutive nasopharyngeal swab samples received between June and July 2020 by the laboratory with presumptive positive SARS-CoV-2 RNA by cobas® (a positive result for the E gene target but a negative result for the ORF1a gene target). These are specimens with a borderline viral titre close to the assay’s limit of detection (LOD) (100 copies/mL) (3) as indicated by their high Ct values. These samples were re-tested within the same day using the Xpert® Xpress SARS-Cov-2 assay. This assay detects the N2 and E gene targets. Similar to the reporting algorithm of the cobas® test, samples with only detectable E gene by the Xpert® assay are considered as presumptive positive with recommendation for re-testing (Xpert® Xpress SARS-Cov-2 assay package insert). The LOD of Xpert® assay was determined by Wolters et al. to be 8.26 copies/mL (4).
Of 24 presumptive positive samples by cobas test, 17 had Ct values generated for both E and N2 gene targets, 2 had Ct value for E gene target only, 4 had Ct value for N2 gene target only, and 1 was negative for both E and N2 gene targets when they were tested using the Xpert® assay. Close agreement between the Ct values of E gene target of the two assays was observed, however, with a high standard deviation (Figure 1). Stochastic effect, which is a prominent observation due to random variations when examining low amounts of PCR targets (5) could cause fluctuation of Ct values between replicate analyses even within the same assay.
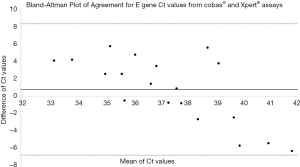
Despite differences in reagent chemistry and cycling parameters of the 2 assays the Ct values for the E gene were quite comparable and the assay that targets SARS-CoV-2 N2 gene is able to detect more cases with low viral load compared to an assay that utilizes the ORF1a region. This adds additional evidence to the observation that the N2 gene persists longer than fragments from the ORF1a (2).The use of more than one molecular kit and the use of assays with more than one viral gene target can maintain high sensitivity for detection of RNA viruses with frequent mutations and capability to detect specific viral gene fragments that may persist longer than others.
Acknowledgments
We acknowledge the staff in the Department of Laboratory Medicine, National University Hospital Singapore who contributed to the laboratory diagnostic services for SARS-CoV-2.
Funding: None.
Footnote
Provenance and Peer Review: This article was a standard submission to the journal. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm-21-17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lv DF, Ying QM, Weng YS, et al. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clin Chim Acta 2020;506:172-5. [Crossref] [PubMed]
- Zóka A, Bekő G. Distinct changes in the real-time PCR detectability of certain SARS-CoV-2 target sequences. Clin Chim Acta 2020;507:248-9. [Crossref] [PubMed]
- Broder K, Babiker A, Myers C, et al. Test Agreement between Roche Cobas 6800 and Cepheid GeneXpert Xpress SARS-CoV-2 Assays at High Cycle Threshold Ranges. J Clin Microbiol 2020;58:e01187-20. [Crossref] [PubMed]
- Wolters F, van de Bovenkamp J, van den Bosch B, et al. Multi-center evaluation of cepheid xpert® xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol 2020;128:104426 [Crossref] [PubMed]
- Weusten J, Herbergs J. A stochastic model of the processes in PCR based amplification of STR DNA in forensic applications. Forensic Sci Int Genet 2012;6:17-25. [Crossref] [PubMed]
Cite this article as: Poon KS, Tee NWS. Comparing the performance of two commercial assays for detection of SARS-CoV-2 in samples with low viral load. J Lab Precis Med 2021;6:23.


