A clinical laboratorian’s journey in developing a machine learning algorithm to assist in testing utilization and stewardship
Highlight box
Key findings
• ML DT algorithm developed appears to provide an opportunity to rule out patients with suspected TTP in resource limited clinical laboratories.
What is known and what is new?
• A pretest probability tool currently available to evaluate TTP is the PLASMIC score.
• AS13 activity assay is required to confirm TTP.
• The AS13 assay is highly complex and overutilized.
• Utilizing a ML DT algorithm as decision support tool in potential TTP cases.
What is the implication and, and what should change now?
• The use of ML/AI algorithms provides opportunities to improve testing utilization and guiding physicians in diagnostic dilemmas. As laboratory specialists become more involved in AI/ML initiatives, institutions will need to provide them with a modern IT infrastructure with adequate resources to enable these efforts.
Introduction
Background
The potential future role of the laboratorian and machine learning (ML)
ML has the potential to be an excellent tool in the clinical laboratory to improve workflow efficiency and testing utilization (1-3). Broadly, artificial intelligence (AI) is defined as the ability of computers to emulate human thinking. ML, a subcategory of AI, uses algorithms to systemically learn and recognize patterns from data with the capacity to improve upon its analysis (1). For most laboratory specialists, the concepts of ML are nebulous. Typically, laboratory specialists have little to no experience in data processing, programming, and ML techniques. Further, while clinical informatics fellowships have only recently started incorporating basic ML concepts and data utilization techniques within their curriculum (4), current pathology trainees (both residents and fellows) mostly are not exposed to general pathology informatics content in their training (5).
Given the certainty that future ML algorithms will continue to be integrated within clinical laboratory technologies, laboratory specialists will necessarily need to be involved with their implementation, utilization, and support (1,2,6). Notably, professional laboratory specialists are responsible for all diagnostic technologies and instrumentation in the lab, including their verification, validation, implementation, and maintenance. Clinically, this responsibility extends to the patient results themselves, with pathologists and other laboratory specialists providing interpretation and consultation services—intimate knowledge of all post-testing calculations and algorithmic modifications is fundamental to high quality laboratories. Thus, the question is not if the laboratorian will be involved with ML/AI, but instead to what extent that involvement will be.
A guide in ML for the clinical laboratorian by a clinical laboratorian
Currently, laboratory specialists who want to learn ML must make a substantial independent effort to teach themselves and find resources outside of their local resource setting (7-9). This can involve a significant expenditure of time and energy, all of which is uncompensated due to its independent nature and lack of direct institutional support. Consequently, the laboratorian’s effort to learn ML comes at his or her own expense, with many laboratory specialists discouraged against learning even basic ML/AI techniques. However, it is crucial that laboratory specialists continue to learn about new technologies in order to improve and push forward the laboratory of the future (1,10). This article is dedicated to that effort, documenting the journey of a clinical laboratorian, inexperienced in ML, programming, and advanced data analytics to develop a ML process for the clinical laboratory. The hope is that this will inspire clinical laboratory specialists and serve as both a roadmap and call to action for others to begin the process of acquiring ML/AI knowledge so they too can help drive their own laboratory into the future.
The current state of the laboratory and diagnostic test utilization
Although there has been a significant national clinical laboratory staff shortage for some time, the COVID-19 pandemic accelerated this trend and the impact is widespread and profound (11-17). The staff shortage impact is felt in almost every area of the lab, including decreased economic resources, overly complex workflow processes, increased result turnaround times, and worsening staff burnout/turnover. Despite these difficulties, the laboratories have continued to make significant efforts to maintain excellent quality in diagnostic testing by implementing more automation, enhancing workflow processes, enacting programs to recruit and retain staff, improving staff productivity, and reducing overly complex assays (18-23).
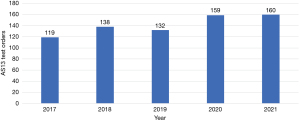
Compounding the current state of the laboratory is inappropriate test utilization that, while not a recent phenomenon, continues to be a significant laboratory (and healthcare) burden (24-31). Specifically, the focus here is test overutilization. Overutilization occurs for a variety of reasons, including ordering provider underestimation of overutilization, lack of understanding of costs, diagnostic uncertainty, unawareness of laboratory resources, habitual practice, and institutional culture (24-31). As one can imagine, the combination of worsening staff shortages and inappropriate diagnostic test utilization is unsustainable (32-34). Similarly, just as the COVID-19 pandemic exacerbated the staffing shortage, it also compounded inappropriate utilization issues, which in our case was especially notable with coagulation testing (35). In our institution, we have an ADAMTS13 (AS13) assay that, at baseline, is likely overutilized and increased during the height of COVID-19 pandemic years (Figure 1). Here, we seek to find a unique solution to inappropriate AS13 testing by developing a ML algorithm with the intention of implementing into the clinical workflow in the future. Before discussing the ML algorithm development, a brief review of the basics of AS13 testing and the role it has in thrombotic thrombocytopenic purpura (TTP) will follow for context.
The physiological role of AS13 in hemostasis
The von Willebrand factor (vWF) molecule is a multimeric glycoprotein synthesized and stored by megakaryocytes and endothelial cells, with platelets also storing vWF in their alpha granules. Importantly for our discussion, vWF is secreted by both platelets and endothelial cells and is essential for primary hemostasis (36). Primary hemostasis is the process of platelet clot formation in the setting of injury to blood vessels. Appropriate primary hemostasis requires that platelet adhesion and aggregation occur at the site of injury and vWF plays a significant role in both of these important processes (36) (Figure 2).
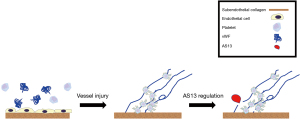
Currently, the only known role of AS13 (a disintegrase and metalloproteinase with a thrombospondin type 1 motif, member 13) is to regulate vWF (37). AS13 is a protease that cleaves vWF into smaller molecules (36). The significance of cleaving vWF into smaller molecules is that it reduces vWF capacity to induce thrombosis; thus, participating in the regulation of both clot formation and extension during primary hemostasis (36,37) (Figure 2).
AS13 and TTP
TTP is a rare thrombotic microangiopathy (TMA) characterized by severe thrombocytopenia, microangiopathic hemolytic anemia, and microvascular platelet-rich thrombi leading to end organ ischemic injury. TTP can be categorized as either acquired or inherited. The inherited form of TTP, also called Upshaw-Schulman syndrome, represents approximately 5% of the total TTP cases (38). It is caused by a genetic alteration of the AS13 gene that renders the protease ineffective in cleaving vWF (37). However, given that acquired TTP represents the majority of TTP cases (~95%), it will be the focus of this article (38).
The approximate annual incidence of TTP is 1–6 cases per million/year (37,38). Acquired TTP is caused by an autoimmune mechanism (antibody inhibitor) that leads to significantly reduced activity levels of AS13 (37,38). The decreased activity levels of AS13 results in excessive “sticky” large (or ultra-large) vWF multimers that promote platelet microthrombi formation. The combination of decreased AS13 activity and excessive large vWF is the pathological mechanism of TTP (36,37).
In our laboratory, the AS13 assay (The Immucor Lifecodes ATS-13® assay, lmmucor GTI Diagnostics Inc., Waukesha, WI, USA) measures the patient’s AS13 activity by providing a reagent substrate that is a synthetic fragment of the vWF molecule containing the AS13 cleavage site. When the synthetic molecule is cleaved by the patient’s AS13 in the sample, fluorescence is released from the molecule via fluorescence resonance energy transfer (FRET) technology. The fluorescence is then measured, which is then compared to a calibration curve that derives the AS13 activity level. A reflex testing protocol is initiated if the AS13 activity level is <20%. If AS13 activity is reduced due to autoantibodies, the reflex test protocol provides the relative amount of activity inhibition by the anti-AS13 antibodies (expressed in Bethesda units). The discrepancy between the potential diagnosis of TTP of <10% activity (see below for details) and our reflex testing at <20% has to do with laboratory experience and limitations of the assay rather than a necessary valid diagnosis of TTP between 10–20%. In our experience, the FRET assay occasionally yields AS13 activities of 10–13% (rarely up to 15%) in cases of definite TTP; analysis of the same specimen by enzyme-linked immunosorbent assay (ELISA) assay typically will read as <10%. The latter topic is beyond the scope of this article and we will continue our discussion with AS13 activity levels <10% for the diagnosis of TTP.
The importance of AS13 testing and pretest probability
An AS13 activity level of <10% is required to confirm the diagnosis of TTP (39). An activity level of >10% tentatively rules out TTP depending on the level above 10% and clinical context; Thus, leaving the differential to other potential TMA and TMA-like syndromes, such as hemolytic uremic syndrome, complement mediated TMA, transplant associated TMA, disseminated intravascular coagulation, and COVID-19 associated coagulopathy (37,38,40). Ideally, once the diagnosis of TTP is confirmed, treatment can be swiftly initiated. Therapeutic plasma exchange (TPE), in conjunction with corticosteroids, is the mainstay of treatment (41) and removes the antibody inhibitor from the plasma that is affecting the AS13 activity. Before TPE, the mortality rate of TTP was >90% (37,42). TPE removes the casual antibody from the patient’s plasma while simultaneously replenishing AS13. The advent of TPE in the treatment of TTP reduced mortality from >90% to 10–20% (37,43), with other adjunctive therapies for TTP available (caplacizumab, rituximab, cyclosporine, and others) (41,44,45).
Despite the importance of diagnostic testing for TTP, many hospitals do not have access to AS13 testing onsite. In fact, most hospitals send the AS13 test out to a reference laboratory, inevitably leading to a delay in results. With this in mind, the guidelines from the International Society on Hemostasis and Thrombosis (ISTH) for the diagnosis of TTP have embedded three recommended testing scenarios when testing is available either within 72 h, >72 h but <7 days, or not available (38). The conclusion is TTP is a medical emergency and therapy should be started based on clinical presentation and level of suspicion (37,38). However, it is important to remember that for patients presenting with a suspected TMA, TPE is first line treatment for TTP but may not be effective with other TMAs. Therefore, it is important for physicians to be able to distinguish between conditions to avoid unnecessary, inappropriate and or delayed treatment.
Given the importance of AS13 testing for accurate diagnosis and treatment of TTP, why don’t most labs have onsite testing? Although each institution may have individualized reasons for this, most will fall under a combination of limited economic and staffing resources, complexity of performing the test, and low incidence of the disease (46,47). Thus, many physicians rely on clinical pretest probability assessments to initiate treatment and then wait for the send out test results to return for confirmation (38).
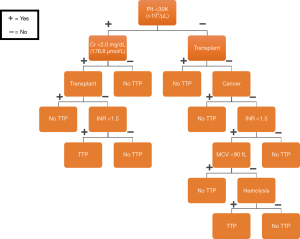
The pre-test probability may be evaluated using different clinical and laboratory parameters. One of the most well-known pre-test probability tools available is the PLASMIC score, which was developed in 2017 (48). The PLASMIC score should only be applied to patients who have both a platelet count (Plt) of <150×103/µL (K) and schistocytes visible on the peripheral blood smear. The PLASMIC score consists of the following seven features: (I) Plt; (II) hemolysis; (III) absence of active cancer; (IV) absence of transplant; (V) mean corpuscular volume; (VI) prothrombin time (PT)-international normalized ratio (INR); and (VII) creatinine (Cr). See Table 1 for details. Each feature is assigned one point and the total score is associated with a risk for TTP, see Table 2.
Table 1
| Feature | Feature description | Assigned point |
|---|---|---|
| Platelets | If <30K† | 1 |
| Hemolysis | If retic count >2.5% or undetectable haptoglobin or indirect bilirubin >2 mg/dL (34.2 µmol/L) | 1 |
| Absence history of transplant | Solid-organ and stem-cell transplant | 1 |
| Absence active cancer | Active cancer is considered if treated within the past year | 1 |
| Mean corpuscular volume | If <90 fL | 1 |
| Creatinine | If <2.0 mg/mL (176.8 µmol/L) | 1 |
| Prothrombin time—INR | If <1.5 | 1 |
†, K: ×103/μL. INR, international normalized ratio.
Table 2
| PLASMIC score total (pts) | Approximate TTP risk (%) |
|---|---|
| 0–4 | <5 |
| 5 | 5–25 |
| 6–7 | 60–80 |
Inhouse testing and the PLASMIC score limitations
In general, when the PLASMIC score is applied correctly, it can be an effective tool for a rapid assessment of a potential TTP case in patients presenting with a TMA (48-54). Additionally, utilizing the PLASMIC score properly could lead to significant cost savings by reducing unnecessary TPEs, decreasing AS13 testing utilization, and lowering subspecialty consultations expenditures (46,47). Unfortunately, application of the PLASMIC score properly does not occur frequently with orders seen at our health system’s institutions, a pattern that is likely the same at other institutions as well. In 2021, we began an initiative to review AS13 testing orders for cancellation (Figure 3). This process included laboratory techs, pathology residents and pathology attendings reviewing patient charts, calculating the PLASMIC scores, consulting with the ordering providers, and adding educational comments to the results. Our investigation revealed that a majority of the orders the lab canceled were due to a combination of patients with Plt >150K, lack of schistocytes on the peripheral blood smear, and hemolysis often explained for other identifiable reasons. Interestingly, review of the medical chart often showed inappropriate PLASMIC score application, if it was used at all (data not published). Our experience suggests the PLASMIC score tends to be overestimated, leading to increased inappropriate test utilization and expending limited laboratory resources.
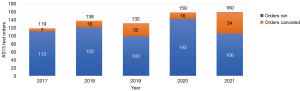
Another observation seen in the data that was indirectly alluded to above is the abrupt increase in testing when the COVID-19 pandemic occurred. One of the reasons for this increased laboratory testing early in the pandemic was due to CAC (40,55). Although research in CAC points to it likely being a distinct entity in which TTP testing was often pursued as a rule out diagnosis due to an overlap between clinical presentation and laboratory test results (35,40). However, the research shows that AS13 activity is not depleted to levels seen in TTP (40,55). In general, concerns for CAC led to an increase with all coagulation testing, including AS13 testing during the pandemic (see years 2020 and 2021 in Figure 1) (35). Finally, although our laboratory is moving to a semiautomated assay, it continues to be a complex test requiring manual processes and expertise in the interpretation of results. Despite some of the automated improvements to the assay, it is important to remain vigilant and continue to improve utilization in diagnostic testing for overall cost effectiveness, efficiency, and quality of patient care (56).
Rationale and knowledge gap
Finding potential solutions around PLASMIC score applicability
As mentioned above, the application of the PLASMIC score in a clinical setting is often inappropriately performed, leading to overestimates in the total PLASMIC score. This therefore favors the inappropriate ordering, and increased utilization of AS13 testing which also has the potential to inappropriate management and treatment. The negative predictive value (NPV) of the PLASMIC score could be increased by varying the weights of the PLASMIC score features based on the clinical circumstance. While one of the original articles in the development of the PLASMIC score did show the odds ratio for each feature, the emphasis remained on the using total score for risk assessment (48). In contrast, is there an optimal combination that may involve an algorithmic application of the PLASMIC score features instead of using the most important features in a sequential manner to identify the prime NPV?
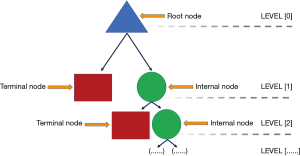
In general, algorithmic flowcharts such as a decision tree (DT) are widely used in medicine across disciplines. DTs are often used as a clinical decision support (CDS) tool to assist in the diagnosis and management of various conditions in medicine (57). A DT has a hierarchical flowchart structure where features are located at decision nodes that guide users along a path to the next node until the final outcome is reached. Formally, the root node is the starting point on the DT leading to all other sub nodes. Internal nodes are bifurcation points directing users to either other internal nodes or a terminal node. A terminal node is a final outcome of the DT, see Figure 4.
In our experience, most established DT algorithms in medicine are developed by a combination of literature review and expert opinion. However, while these algorithms can be sufficiently accurate for their intended use cases, they are not developed in a standardized robust manner. They usually require multiple physician expert time and are often limited to scenarios that are relatively not overly complex (58-61). Recently, ML developed algorithms have gained traction in medicine and their use in CDS tools is noted in a number of areas, including laboratory medicine (62-65).
Identifying the “right” ML algorithm by a novice
ML algorithms can be difficult to understand or obscure to the everyday laboratorian or physician. Many algorithms appear opaque and perplexing due to their mathematical complexity and inability to provide “palatable” visualization of how decisions and predictions are developed and executed. Further, strong arguments can be made against using obscure and opaque algorithms, sometimes called “black box” algorithms, in medicine as they may potentially border along unethical boundaries (65,66). “Black box” AI/ML algorithms are often a combination of an individual’s lack of understanding of how predictions are made, complexity of a model in which a simple explanation/representation cannot be crafted such as deep learning systems, and potential propriety algorithms which inherently lead to a lack of transparency. However, using ML DT algorithms may avoid many of these potential problems because these algorithms can be visualized and inspected for interpretability in the predictions that are made (3). Further, many healthcare providers are familiar with DT algorithms as discussed above.
Objective
If a PubMed advanced keyword search of “machine learning” and “thrombotic thrombocytopenic purpura” is performed, only two articles are retrieved from the literature (67,68). The intention of the first study is to predict TTP and other TMAs using ML, with it considering the difficulty of onsite AS13 testing and the unreliability of the PLASMIC score regarding ethnicity for the TMA differential (67). Additionally, it explores the use of an ensemble ML method to address this issue by evaluating nine ML algorithms, including a DT. The group highlights multiple features, some of which are from the PLASMIC score, but did emphasize that the DT algorithm was the least predictive of the ML methods. The second study focused on identifying a biomarker for the prognosis of TTP, using ML to identify D-dimer as the strongest predictor of the prognosis, mortality, and thrombotic events in TTP (68). In contrast to both of these studies, the objective of our study is distinct in that we are only using the PLASMIC score features to develop a DT algorithm that is optimized for a high NPV. We present this article in accordance with the STARD reporting checklist (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-23-11/rc).
Methods
Finding the right software and programming language
As a novice with absolutely no experience, identifying the appropriate software and programming language to develop a ML DT type algorithm was crucial. This process involved a combination of criteria, including finding easy-to-use software, an abundance of free learning resources, ease of programming language, DT capability, and finally, cost (Figure 5). Based on these criteria, PythonTM was selected for the programming language. Anaconda®, an open source software, was used to manage projects, which included removing and installing statistical algorithms and visualization packages for the ML model development.
To approach the question as to whether we could discover an optimal sequence of the PLASMIC score features, we had to find which feature provided the most information for each DT node development that separated the patients with TTP from those without. As discussed above, that arrangement may or may not have been the feature with the greatest odds ratio. A quick calculation of all the possible arrangements (or permutations) of the seven features revealed 5040 (7! = 5040) potential sequences. The next step then involved each arrangement being tested against the training data to isolate the highest NPV. Of note, the binary element (yes or no) of each feature was not considered in the latter arrangement calculation. Overall, this was not a simple task easily tackled using simple flat-file spreadsheets—we concluded that the use of ML and other mathematical tools was the best approach for addressing our problem’s complexity.
In order to optimize the conditions of a ML DT for a high NPV, we chose the mathematical approach of first objectively measuring information regarding each feature used in the algorithm and then applying those measurements to resolve the proper sequence of features. This approach is based on Information Theory, specifically using Shannon entropy as a way to measure information (69). Entropy, in simple terms for our discussion, measures the amount of “mixing” in a dataset, which in our case signifies patients with vs. without TTP (non-TTP). For example, if our data set contains 20 patients with a 10-10 mix of TTP vs. non-TTP, then it has maximum entropy due to the fact that if we were to randomly select a patient from our data pool, we would have an equal probability of retrieving either a TTP or a non-TTP patient. However, as new information is introduced through the application of features, then the dataset mix shifts away from being equal (10-10 mix) and we are able to classify TTP and non-TTP into purer sub datasets. Hypothetically, each feature applied should inherently shift the original dataset in such a way that entropy would decrease, as seen in Figure 6A,6B. Overall, the goal of the DT was to shift our dataset from high to low entropy in order to develop a model that could more accurately predict TTP vs. non-TTP.

Data collection
This retrospective observation study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), after receiving approval from the University of Florida Institutional Review Board (IRB #202202290 October 11, 2022). The data collection was based on information available in the electronic medical health record. This protocol did not interfere with any standard of care practice as it was retrospective. Thus, individual informed consent was waived.
A hybrid approach, consisting of both inhouse and literature data, was taken to acquire the data to train and test the DT ML model. Our inhouse data consists of the years 2017 until 2021. Our data identified 14 newly diagnosed cases of TTP, for which the PLASMIC score and AS13 level were collected for each of these patients from the electronic medical record. We also randomly selected 16 control patients for which the PLASMIC score was collected and the AS13 activity levels were present, ranging between 12–30%. AS13 activities between 10–15% were checked with an alternative assay method to confirm our results by an independent laboratory outside of our health system.
Due to the limitations with our inhouse data for newly diagnosed TTP patients (low volume, n=14), we decided to supplement this dataset using the literature. We used specific criteria to collect consistent and appropriate data per patient for our study from the literature, including: (I) adult population; (II) PLASMIC score containing individual patient feature scores; (III) AS13 activity level prior to treatment present; (IV) new diagnosis of TTP. Three articles met the criteria for our study (49,50,70). In toto, the combined inhouse and literature dataset consisted of 104 patients (30 inhouse, 74 literature-derived) with an equal mix of TTP to non-TTP patients (52 each).
Optimizing the ML DT with the training data set
The PLASMIC score data was collected for the entire dataset (inhouse and literature) and entered into a spreadsheet. Each patient and their respective corresponding PLASMIC score features were coded into binary units (feature present =1, feature absent =0). For example, for the feature ‘platelet count <30K/µL’, all platelet counts <30K/µL were coded as 1 (present) and anything >30K/µL was coded as 0 (absent). All PLASMIC score features were scored similarly and the spreadsheet converted to a CSV file.
The next step for ML algorithm development and statistical analysis involved installation of the required software packages in PythonTM. Key software packages included the DT classifier and train-test-split. The DT classifier package allows for a selection of options to be used in the development of the DT. For our purposes, the entropy option was selected for the analysis of the quality of the node splitting, as discussed above. Additionally, another tool was used to fine tune the DT using the cost complexity pruning (CCP) parameter. See discussion below for details.
The purpose of the train-test-split package was to split all of the data randomly into two subsets, training data (75%) and test data (25%). A supervised ML approach was subsequently employed, where training data is first labeled for appropriate outcomes (TTP and non-TTP) and then used to train the DT algorithm. Test data was held aside until initial model training was complete and fine-tuned, after which it was used to evaluate the trained algorithm’s performance on new unseen data.
Of note, while our goal was for the model to be as accurate as possible in distinguishing between TTP and non-TTP patients, our approach intentionally optimized the negative predictive value more than overall accuracy (i.e., maximum confidence when model output equals non-TTP). While this invariably translates to potentially increased false positives results (outcomes classified as TTP are actually non-TTP), our overall goal was not to restrict testing on any potentially true TTP cases.
In supervised ML with DTs, a well-known limitation is the propensity to overfit the data during training. Qualitatively, this is apparent by observing the large size (number of nodes and levels) of the DT. With overfitting, DT algorithms give accurate predictions for the training data, but then do not necessarily perform well on the test (new) data. As an analogy, a student (DT) has learned to get all of the answers correct (outcome label: TTP & non-TTP) on a practice exam (training data) given by the professor. The student (DT) has made such an effort to get all of the answers correct on the practice exam that they essentially memorized the specific practice test questions and answers rather than learning the concepts from the practice exam. Thus, when the real exam (new unseen data) is given, the student (DT) performs poorly.
When using the DT classification package, overfitting naturally occurs as the package scripts prioritize training (creating as many DT nodes, or branches to fit the data) over generalizability (using the minimum number of nodes to get the best accuracy of the model across all data). Additional functions are therefore included in the package to help ‘prune’ the weakest branches and minimize overfitting of a DT algorithm, typically by: (I) requiring a minimum number of samples in a node before considering a new split, and (II) limiting the number of internal nodes in any given DT. For our purposes, we made use of the CCP parameter and cross validation (CV) protocol to help minimize overfitting of our DT.
With the CCP, a complexity parameter (alpha) is calculated for each node of the tree, which then allows for setting an overall alpha value that can act as a limit, or tuning parameter, to prune the “weakest” branches of the DT. As the CCP alpha increases, the nodes with the smallest alphas (“weakest” branches) are pruned first, however with high alphas, more and more of the tree gets pruned. In the end, the goal of the CCP alpha process is to define the alpha parameter such that training accuracy only slightly decreases while testing accuracy increases. In essence, this allows for fine tuning of the DT while minimizing the error rate and increasing data generalizability.
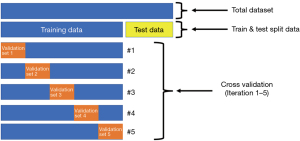
The second function used was the CV protocol. The CV performs 5 iterations of running the training dataset to train the DT model. Importantly, before each iteration a subset of the training data, or validation set, is removed and hidden until the training is completed for that iteration. At this point, the validation set is then tested on the trained model and accuracy scores are calculated. Each iteration validation set is different from the previous set. Thus, the model was trained and “validated” on different combinations of the training data to assess generalizability by evaluating consistency of the accuracy score for each iteration (Figure 7).
Results
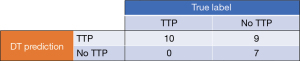
For our DT model, the initial accuracy was ~94%, post training but without fine tuning using the CCP or CV functions, for distinguishing TTP vs. non-TTP patients. Notably, the preliminary DT was large and complex, with a total of 7 levels and 43 nodes. As expected, however, when evaluated against the test dataset, the DT model performed much lower at ~65% overall accuracy. Despite this initial lower overall accuracy with new data, the model did accurately predict non-TTP patients in the test set, with a 100% negative predictive value (Figure 8).
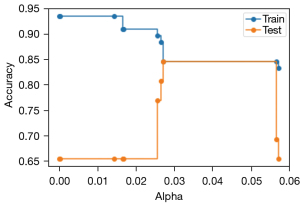
Next, the DT model was fine-tuned using the CCP alpha pruning process to achieve better overall accuracy with new data while also maintaining its excellent NPV. The accuracy of the model on both the training and test dataset was measured for every increase in the alpha parameter, with the results plotted to visualize the differences in model performance (Figure 9). With no fine tuning (alpha =0), baseline accuracy of the model was 94% using the training data and 65% using the test data. With increasing alpha parameter values, the number of model nodes decrease, resulting in improved accuracy on the test data but lowered accuracy on the training data. As seen in Figure 9, at an alpha between 0.025 and 0.03, the DT model’s training and test data performance intersect at 85% accuracy; at this point, increasing the alpha parameter does not change the overall accuracy performance of the model for either dataset. However, when the alpha value approaches 0.06, the performance of the DT model sharply decreases for the test dataset.
Following fine tuning with the CCP function, we then moved to further stabilize and provide additional consistency to our model through the application of the CV protocol. In this situation, a CCP alpha of 0.03 and a CV of five iterations were selected. After each iteration, the DT was trained and the accuracy recorded from the evaluation of the validation data set. The accuracy results for each iteration, respectively, were 81%, 81%, 81%, 73% and 87%. These results suggested the DT performance with the selected alpha was a function of the way the training data and validation set was split for each iteration. Although the accuracy was, to a certain extent, relatively consistent, we still wanted to further refine the model more for use with external datasets by identifying a more ideal alpha that maximizes NPV vs. model accuracy.
To better select a less sensitive alpha to the validation data splits, we used a similar approach to CV protocol, however this time the CCP alpha function was nested within the CV function. By doing this, we were then able to measure the accuracy at different alpha values for each iteration of the CV. From this data, a CCP alpha of 0.0265 was identified that optimized the DT model for maximum generalizability and NPV.
The final DT model, with the alpha set at 0.0265, was then trained with the original training data, followed by evaluation with the original test data. Both training and test overall performance was 81% with this model, with the NPV remaining at 100%, see Figure 10. Comparing the performance and characteristics of the final (pruned) DT against the initial DT model, see Table 3. For reference, the drawn final DT model is shown in Figure 11.
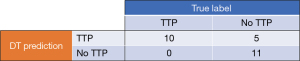
Table 3
| Evaluation characteristic | Preliminary DT | Pruned DT |
|---|---|---|
| Sensitivity (%) | 100 | 100 |
| Specificity (%) | 44 | 69 |
| PPV (%) | 53 | 67 |
| NPV (%) | 100 | 100 |
| Overall accuracy (%) | 65 | 81 |
| DT levels | 7 | 6 |
| DT Nodes | 43 | 19 |
DT, decision tree; PPV, positive predictive value; NPV, negative predictive value.
Discussion
Key findings
Our TTP DT model was able to meet both our functional specifications and primary clinical use case, which was to assist in the appropriate use of our inhouse AS13 assay by providing a high NPV on the testing data set. By insisting on having a high NPV for our model, we can now be confident of negative for TTP outputs from any given patient’s PLASMIC score inputs. Further, we are now able to redirect inappropriate AS13 testing requests, in addition to aiding our clinical colleagues when faced with a potential TMA diagnostic dilemma. Of note, we were also able to optimize the positive predictive value (PPV) of the preliminary DT model as compared to the final ‘pruned’ model, from 53% to 67%, respectively. This PPV is acceptable for our intended purposes.
Strengths, limitations and comparison to similar research
In order to focus on developing a ML algorithm for the first time in our laboratory, we needed to operate from a position that allowed us to use features already relatively established for the diagnosis TTP. We decided to use features from the PLASMIC score as this has been supported in the literature. Second, we wanted to avoid going down the path of using overly complex methods, such as those requiring data transformation/preprocessing, custom programming, and complex ML algorithms like convolutional neural networks. Based on these requirements, we selected to move forward with a DT algorithm given its inherent transparency and ability to be visually displayed in a manner easily understood by medical professionals, regardless of any prior ML knowledge. If successful, we feel this would allow us to implement the algorithm into practice with less barriers.
Despite the strengths to our approach, there are limitations of our model which include the issue of overfitting of the DT algorithm and our use of a relatively small dataset for model development. We tried to ameliorate overfitting by using additional functions such as CCP and CV, during model development. Another inherent limitation of a DT is that it can also be unstable with small variations in training data sets, thus generating very different trees (71). We utilized the CV function to give us some insight to this instability, however, we remain concerned about this potential. There are additional techniques one can use to lower potential variance, such as bagging or boosting methods. We preliminarily used a random forest bagging method for our model evaluation (data not shown) and to date we feel that the DT NPV is adequate. Furthermore, it is important to highlight that the calculated negative predictive value is only true for the prevalence of TTP within the evaluated cohort, which is 50%. This does not represent the true prevalence of disease in the general population (~0.0003%) nor the true prevalence in our laboratory based on testing with AT13 (~2.5%). Assuming that the DT model would preserve the current sensitivity (100%) and specificity (69%) with further testing in datasets with decreasing prevalence such as 30%, 10% or 0.5%, the NPV should not significantly change and remain approximately 100%. However, false positives will increase but the rate of increase diminishes as the prevalence continues to decrease. To reiterate, the goal of the model is to have an excellent NPV that should continue to perform adequately in reaching lower levels of prevalence. Nevertheless, further testing is absolutely required and we are currently gathering a more robust dataset to fully validate the model. Additionally, a direct performance comparison between the ML DT algorithm and PLASMIC score should be addressed, specifically comparing the NPV.
The limited available dataset we encountered is a concern, however given the frequency of the disease, it’s not surprising only a limited number of cases could be identified, even after using data from the literature to supplement our smaller inhouse dataset. Fortunately, a DT algorithm might perform adequately despite a limited dataset as long as the features number is also relatively minimal (72). Given these limitations, we look forward to testing the our TTP model on an external validation dataset. If the performance is acceptable, we would proceed to test the model in parallel to our current clinical workflows so we can better note discrepancies and determine whether additional fine tuning is required.
As mentioned in the introduction section, to our knowledge, there is currently very limited research in the area of TTP and ML. At this time, there were only two studies that evaluated TTP with ML techniques (67,68). Again, the approach to both of these studies are different from our study in both selected features and ML algorithms utilized.
Implications and actions needed
In this study, we used PLASMIC score features to develop a supervised ML DT model to rule out TTP. Our primary intention for developing this model was to assist our laboratory with future test utilization and stewardship of the AS13 assay. Secondarily, we wanted to demonstrate how a laboratorian inexperienced in ML could acquire the skills necessary to develop a ML model, including basic concepts in supervised ML such as DT analysis, overfitting of training data, and fine tuning of model performance to improve generalizability to unseen data.
When developing and deploying ML models, there are many different aspects to consider and steps to complete prior to letting them loose in the wild. In fact, for many in healthcare, the ML process tends to stop after the proof of principle/concept phases given the inherent complexity and burden of getting a clinically oriented ML model ready for use within healthcare information technology (IT) infrastructures. Recently, Lavin et al. published a system engineering approach to ML and AI systems using technology readiness levels (TRL) (73). TRLs provide users interested in developing ML algorithms a robust framework to follow, recognizing that algorithms don’t exist in a vacuum and instead are composed of many different components and connected systems. Further, the TRL approach considers the maturity of ML models and systems, including their accompanying data pipelines and software dependencies. Overall, TRLs range in maturity from TRL0 (first principles) to TRL9 (deployment).
Currently, our TTP ML model is in its earliest stages and we would classify it at a TRL2, or proof of principle, maturity level. At a TRL2, we have successfully developed and run a model in a simulated clinical/research environment, using data similar to that to which will be used in clinical practice. Going forward, we plan to engage our institutional IT group to determine the next best steps for clinical deployment. Of note, while we could move systematically through each of the TRL levels to create our own ML system, we recognize that the effort involved in doing so is beyond the scope of typical clinical laboratory practice. Instead, by partnering with our IT colleagues, we will look to leverage existing IT systems, policy, and procedures in order to better achieve our goals.
Concomitant to IT systems and integration evaluation, we will also begin efforts to assess our model’s robustness by exposing it to a wider variety of real-world data. These efforts will include steps taken to assess our model’s bias, ensure a proper ethics review has been performed, and review potential effects of our model on both laboratory and clinician decision making. We will also engage our institutional compliance office to assess our model’s regulatory status and whether it qualifies a medical device per the recent Clinical Decision Support Software guidance released by the Food and Drug Administration (FDA) (74). Finally, once proper data pipelines are established, we plan to “shadow” test the model in the clinical environment by creating a parallel testing workflow using actual patient cases coming through our laboratory.
We should note that even if one is able to perform all of the steps as described above, there are still additional considerations that must be managed prior to deployment of our model. For example, with low enough volumes, it may be entirely realistic to manually implement our model on local workstations in the lab vs. investing in a costlier ML platform. Additionally, we will need to consider whether the algorithm will be used solely within the laboratory to manage AS13 testing vs. releasing the predictions from the model to the electronic health record (EHR) and allowing clinicians to act on the information. Another option for us could be to leverage our EHR’s AI/ML module for model deployment, however that would require our project to go through institutional prioritization and demand review. From a regulatory perspective, we will also need to review whether our model would qualify as a medical device vs. a CDS tool per the Federal FDA Software as a Medical Device (SaMD) guidelines (75). This decision will hinge on multiple factors, including the intended use of the model, the clinical data inputs, and how the model outputs will be used by pathologists, laboratory specialists, and/or other health care professionals when making specific patient care decisions. Fundamentally, there are many different deployment issues for us to consider as we move forward.
Conclusions
Overall, developing a ML DT model provided an excellent opportunity to immerse ourselves in ML techniques and programming. We are not yet experts in this area, but we believe we have made great strides moving toward it and preparing ourselves for the clinical laboratory of the future. More importantly, we want to develop a framework for implementing ML-based decision support tools developed by clinical laboratory specialists within our institution. It does have advantages. It is important to note that both conventional and advanced statistical methods/models may be appropriate to utilize in many situations in the clinical laboratory and may offer advantages over ML models. However, this analysis, while important, is beyond the scope of this current article. Nevertheless, this will create opportunities for laboratory specialists to be stewards of diagnostic testing. The difficulty for everyday laboratory specialists to acquire and develop ML techniques is due to lack of training, limited experience, and limited institutional support. The healthcare system and physicians rely heavily on laboratory services to aid in clinical decisions, with the laboratory often at the forefront of technology implementation in healthcare—we envision clinical laboratories further being at the forefront of AI and ML. Finally, as laboratory specialists become more involved in AI/ML initiatives, institutions will need to provide them with a modern IT infrastructure with adequate resources to enable these efforts.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Lee Schroeder) for the series “Data-Driven Laboratory Stewardship” published in Journal of Laboratory and Precision Medicine. The article has undergone external peer review.
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-23-11/rc
Data Sharing Statement: Available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-23-11/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-23-11/coif). The series “Data-Driven Laboratory Stewardship” was commissioned by the editorial office without any funding or sponsorship. NSH reports honoraria for a presentation given at the American Association for Clinical Chemistry (AACC), 2021 annual meeting and support for attending and travel to the College of American Pathologist (CAP), 2022 meeting. MJM reports honoraria for a presentation given at the AACC annual meeting, 2022. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the University of Florida Institutional Review Board (IRB #202202290 October 11, 2022). The individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haymond S, McCudden C. Rise of the Machines: Artificial Intelligence and the Clinical Laboratory. J Appl Lab Med 2021;6:1640-54. [Crossref] [PubMed]
- Herman DS, Rhoads DD, Schulz WL, et al. Artificial Intelligence and Mapping a New Direction in Laboratory Medicine: A Review. Clin Chem 2021;67:1466-82. [Crossref] [PubMed]
- Punchoo R, Bhoora S, Pillay N. Applications of machine learning in the chemical pathology laboratory. J Clin Pathol 2021;74:435-42. [Crossref] [PubMed]
- Silverman HD, Steen EB, Carpenito JN, et al. Domains, tasks, and knowledge for clinical informatics subspecialty practice: results of a practice analysis. J Am Med Inform Assoc 2019;26:586-93. [Crossref] [PubMed]
- Black-Schaffer WS, Robboy SJ, Gross DJ, et al. Evidence-Based Alignment of Pathology Residency With Practice II: Findings and Implications. Acad Pathol 2021;8:23742895211002816. [Crossref] [PubMed]
- Marin MJ, Van Wijk XMR, Durant TJS. Machine Learning in Healthcare: Mapping a Path to Title 21. Clin Chem 2022;68:609-10. [Crossref] [PubMed]
- Cheng JY, Abel JT, Balis UGJ, et al. Challenges in the Development, Deployment, and Regulation of Artificial Intelligence in Anatomic Pathology. Am J Pathol 2021;191:1684-92. [Crossref] [PubMed]
- Harrison JH, Gilbertson JR, Hanna MG, et al. Introduction to Artificial Intelligence and Machine Learning for Pathology. Arch Pathol Lab Med 2021;145:1228-54. [Crossref] [PubMed]
- Yao K, Singh A, Sridhar K, et al. Artificial Intelligence in Pathology: A Simple and Practical Guide. Adv Anat Pathol 2020;27:385-93. [Crossref] [PubMed]
- Blanckaert N. Clinical pathology services: remapping our strategic itinerary. Clin Chem Lab Med 2010;48:919-25. [Crossref] [PubMed]
- Halstead DC, Sautter RL. A Literature Review on How We Can Address Medical Laboratory Scientist Staffing Shortages. Lab Med 2023;54:e31-6. [Crossref] [PubMed]
- Huq Ronny FM, Sherpa T, Choesang T, et al. Looking into the Laboratory Staffing Issues that Affected Ambulatory Care Clinical Laboratory Operations during the COVID-19 Pandemic. Lab Med 2023;54:e114-6. [Crossref] [PubMed]
- Leber AL, Peterson E, Dien Bard J, et al. The Hidden Crisis in the Times of COVID-19: Critical Shortages of Medical Laboratory Professionals in Clinical Microbiology. J Clin Microbiol 2022;60:e0024122. [Crossref] [PubMed]
- Garcia E, Kundu I, Kelly M, et al. The American Society for Clinical Pathology 2020 Vacancy Survey of Medical Laboratories in the United States. Am J Clin Pathol 2022;157:874-89. [Crossref] [PubMed]
- Garcia E, Ali AM, Soles RM, et al. The American Society for Clinical Pathology’s 2014 vacancy survey of medical laboratories in the United States. Am J Clin Pathol 2015;144:432-43. [Crossref] [PubMed]
- Garcia E, Kundu I, Kelly M, et al. The American Society for Clinical Pathology’s 2018 Vacancy Survey of Medical Laboratories in the United States. Am J Clin Pathol 2019;152:155-68. [Crossref] [PubMed]
- Garcia E, Kundu I, Fong K. American Society for Clinical Pathology’s 2019 Wage Survey of Medical Laboratories in the United States. Am J Clin Pathol 2021;155:649-73. [Crossref] [PubMed]
- Bailey AL, Ledeboer N, Burnham CD. Clinical Microbiology Is Growing Up: The Total Laboratory Automation Revolution. Clin Chem 2019;65:634-43. [Crossref] [PubMed]
- Ellison TL, Alharbi M, Alkaf M, et al. Implementation of total laboratory automation at a tertiary care hospital in Saudi Arabia: effect on turnaround time and cost efficiency. Ann Saudi Med 2018;38:352-7. [Crossref] [PubMed]
- Gudmundsson GS, Kahn SE, Moran JF. Association of mild transient elevation of troponin I levels with increased mortality and major cardiovascular events in the general patient population. Arch Pathol Lab Med 2005;129:474-80. [Crossref] [PubMed]
- Jones BA, Darcy T, Souers RJ, et al. Staffing benchmarks for clinical laboratories: a College of American Pathologists Q-Probes study of laboratory staffing at 98 institutions. Arch Pathol Lab Med 2012;136:140-7. [Crossref] [PubMed]
- Novis DA, Coulter SN, Blond B, et al. Technical Staffing Ratios. Arch Pathol Lab Med 2022;146:330-40. [Crossref] [PubMed]
- Novis DA, Nelson S, Blond BJ, et al. Laboratory Staff Turnover: A College of American Pathologists Q-Probes Study of 23 Clinical Laboratories. Arch Pathol Lab Med 2020;144:350-5. [Crossref] [PubMed]
- Vrijsen BEL, Naaktgeboren CA, Vos LM, et al. Inappropriate laboratory testing in internal medicine inpatients: Prevalence, causes and interventions. Ann Med Surg (Lond) 2020;51:48-53. [Crossref] [PubMed]
- Cadamuro J, Gaksch M, Wiedemann H, et al. Are laboratory tests always needed? Frequency and causes of laboratory overuse in a hospital setting. Clin Biochem 2018;54:85-91. [Crossref] [PubMed]
- Zhi M, Ding EL, Theisen-Toupal J, et al. The landscape of inappropriate laboratory testing: a 15-year meta-analysis. PLoS One 2013;8:e78962. [Crossref] [PubMed]
- Conroy M, Homsy E, Johns J, et al. Reducing Unnecessary Laboratory Utilization in the Medical ICU: A Fellow-Driven Quality Improvement Initiative. Crit Care Explor 2021;3:e0499. [Crossref] [PubMed]
- Tamburrano A, Vallone D, Carrozza C, et al. Evaluation and cost estimation of laboratory test overuse in 43 commonly ordered parameters through a Computerized Clinical Decision Support System (CCDSS) in a large university hospital. PLoS One 2020;15:e0237159. [Crossref] [PubMed]
- van Walraven C, Naylor CD. Do we know what inappropriate laboratory utilization is? A systematic review of laboratory clinical audits. JAMA 1998;280:550-8. [Crossref] [PubMed]
- Hauser RG, Shirts BH. Do we now know what inappropriate laboratory utilization is? An expanded systematic review of laboratory clinical audits. Am J Clin Pathol 2014;141:774-83. [Crossref] [PubMed]
- Sedrak MS, Patel MS, Ziemba JB, et al. Residents’ self-report on why they order perceived unnecessary inpatient laboratory tests. J Hosp Med 2016;11:869-72. [Crossref] [PubMed]
- Plebani M, Lippi G. Is laboratory medicine a dying profession? Blessed are those who have not seen and yet have believed. Clin Biochem 2010;43:939-41. [Crossref] [PubMed]
- Al Naam YA, Elsafi S, Al Jahdali MH, et al. The Impact of Total Automaton on the Clinical Laboratory Workforce: A Case Study. J Healthc Leadersh 2022;14:55-62. [Crossref] [PubMed]
- Plebani M. Appropriateness in programs for continuous quality improvement in clinical laboratories. Clin Chim Acta 2003;333:131-9. [Crossref] [PubMed]
- Marin MJ, Harris N, Winter W, et al. A Rational Approach to Coagulation Testing. Lab Med 2022;53:349-59. [Crossref] [PubMed]
- Harris NS, Pelletier JP, Marin MJ, et al. Von Willebrand factor and disease: a review for laboratory professionals. Crit Rev Clin Lab Sci 2022;59:241-56. [Crossref] [PubMed]
- Sukumar S, Lämmle B, Cataland SR. Thrombotic Thrombocytopenic Purpura: Pathophysiology, Diagnosis, and Management. J Clin Med 2021;10:536. [Crossref] [PubMed]
- Zheng XL, Vesely SK, Cataland SR, et al. ISTH guidelines for the diagnosis of thrombotic thrombocytopenic purpura. J Thromb Haemost 2020;18:2486-95. [Crossref] [PubMed]
- George JN, Nester CM. Syndromes of thrombotic microangiopathy. N Engl J Med 2014;371:654-66. [Crossref] [PubMed]
- Mei ZW, van Wijk XMR, Pham HP, et al. Role of von Willebrand Factor in COVID-19 Associated Coagulopathy. J Appl Lab Med 2021;6:1305-15. [Crossref] [PubMed]
- Zheng XL, Vesely SK, Cataland SR, et al. ISTH guidelines for treatment of thrombotic thrombocytopenic purpura. J Thromb Haemost 2020;18:2496-502. [Crossref] [PubMed]
- Padmanabhan A, Connelly-Smith L, Aqui N, et al. Guidelines on the Use of Therapeutic Apheresis in Clinical Practice – Evidence-Based Approach from the Writing Committee of the American Society for Apheresis: The Eighth Special Issue. J Clin Apher 2019;34:171-354. [Crossref] [PubMed]
- Rock GA, Shumak KH, Buskard NA, et al. Comparison of plasma exchange with plasma infusion in the treatment of thrombotic thrombocytopenic purpura. Canadian Apheresis Study Group. N Engl J Med 1991;325:393-7. [Crossref] [PubMed]
- Akwaa F, Antun A, Cataland SR. How I treat immune-mediated thrombotic thrombocytopenic purpura after hospital discharge. Blood 2022;140:438-44. [Crossref] [PubMed]
- Scully M, Cataland SR, Peyvandi F, et al. Caplacizumab Treatment for Acquired Thrombotic Thrombocytopenic Purpura. N Engl J Med 2019;380:335-46. [Crossref] [PubMed]
- Upadhyay VA, Geisler BP, Sun L, et al. Utilizing a PLASMIC score-based approach in the management of suspected immune thrombotic thrombocytopenic purpura: a cost minimization analysis within the Harvard TMA Research Collaborative. Br J Haematol 2019;186:490-8. [Crossref] [PubMed]
- Kim CH, Simmons SC, Wattar SF, et al. Potential impact of a delayed ADAMTS13 result in the treatment of thrombotic microangiopathy: an economic analysis. Vox Sang 2020;115:433-42. [Crossref] [PubMed]
- Bendapudi PK, Hurwitz S, Fry A, et al. Derivation and external validation of the PLASMIC score for rapid assessment of adults with thrombotic microangiopathies: a cohort study. Lancet Haematol 2017;4:e157-64. [Crossref] [PubMed]
- Jajosky R, Floyd M, Thompson T, et al. Validation of the PLASMIC score at a University Medical Center. Transfus Apher Sci 2017;56:591-4. [Crossref] [PubMed]
- Oliveira DS, Lima TG, Benevides FLN, et al. Plasmic score applicability for the diagnosis of thrombotic microangiopathy associated with ADAMTS13-acquired deficiency in a developing country. Hematol Transfus Cell Ther 2019;41:119-24. [Crossref] [PubMed]
- Li A, Khalighi PR, Wu Q, et al. External validation of the PLASMIC score: a clinical prediction tool for thrombotic thrombocytopenic purpura diagnosis and treatment. J Thromb Haemost 2018;16:164-9. [Crossref] [PubMed]
- Lee CH, Huang YC, Li SS, et al. Application of PLASMIC Score in Risk Prediction of Thrombotic Thrombocytopenic Purpura: Real-World Experience From a Tertiary Medical Center in Taiwan. Front Med (Lausanne) 2022;9:893273. [Crossref] [PubMed]
- Wynick C, Britto J, Sawler D, et al. Validation of the PLASMIC score for predicting ADAMTS13 activity <10% in patients with suspected thrombotic thrombocytopenic purpura in Alberta, Canada. Thromb Res 2020;196:335-9. [Crossref] [PubMed]
- Paydary K, Banwell E, Tong J, et al. Diagnostic accuracy of the PLASMIC score in patients with suspected thrombotic thrombocytopenic purpura: A systematic review and meta-analysis. Transfusion 2020;60:2047-57. [Crossref] [PubMed]
- Iba T, Levy JH, Connors JM, et al. The unique characteristics of COVID-19 coagulopathy. Crit Care 2020;24:360. [Crossref] [PubMed]
- Dickerson JA, Fletcher AH, Procop G, et al. Transforming Laboratory Utilization Review into Laboratory Stewardship: Guidelines by the PLUGS National Committee for Laboratory Stewardship. J Appl Lab Med 2017;2:259-68. [Crossref] [PubMed]
- Healy PM, Jacobson EJ. Common Medical Diagnoses: An Algorithmic Approach. 4th edition. Philadelphia, PA, USA: Saunders; 2006.
- Jablonski AM, DuPen AR, Ersek M. The use of algorithms in assessing and managing persistent pain in older adults. Am J Nurs 2011;111:34-43; quiz 44-5. [Crossref] [PubMed]
- Proposal for clinical algorithm standards. Society for Medical Decision Making Committee on Standardization of Clinical Algorithms. Med Decis Making 1992;12:149-54. [Crossref] [PubMed]
- Hadorn DC, McCormick K, Diokno A. An annotated algorithm approach to clinical guideline development. JAMA 1992;267:3311-4. [Crossref] [PubMed]
- Siddall PJ, Middleton JW. A proposed algorithm for the management of pain following spinal cord injury. Spinal Cord 2006;44:67-77. [Crossref] [PubMed]
- Xu HA, Maccari B, Guillain H, et al. An End-to-End Natural Language Processing Application for Prediction of Medical Case Coding Complexity: Algorithm Development and Validation. JMIR Med Inform 2023;11:e38150. [Crossref] [PubMed]
- Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol 2019;19:64. [Crossref] [PubMed]
- Rabbani N, Kim GYE, Suarez CJ, et al. Applications of machine learning in routine laboratory medicine: Current state and future directions. Clin Biochem 2022;103:1-7. [Crossref] [PubMed]
- Cabitza F, Banfi G. Machine learning in laboratory medicine: waiting for the flood? Clin Chem Lab Med 2018;56:516-24. [Crossref] [PubMed]
- Watson DS, Krutzinna J, Bruce IN, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ 2019;364:l886. [Crossref] [PubMed]
- Yoon J, Lee S, Sun CH, et al. MED-TMA: A clinical decision support tool for differential diagnosis of TMA with enhanced accuracy using an ensemble method. Thromb Res 2020;193:154-9. [Crossref] [PubMed]
- Wang HX, Han B, Zhao YY, et al. Serum D-dimer as a potential new biomarker for prognosis in patients with thrombotic thrombocytopenic purpura. Medicine (Baltimore) 2020;99:e19563. [Crossref] [PubMed]
- Krause P. Information Theory and Medical Decision Making. Stud Health Technol Inform 2019;263:23-34. [PubMed]
- Moosavi H, Ma Y, Miller MJ, et al. Validation of PLASMIC score: an academic medical center case series (2012-present). Transfusion 2020;60:1536-43. [Crossref] [PubMed]
- Uddin S, Khan A, Hossain ME, et al. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak 2019;19:281. [Crossref] [PubMed]
- Althian A, AlSaeed D, Al-Baity H, et al. Impact of Dataset Size on Classification Performance: An Empirical Evaluation in the Medical Domain. Appl Sci 2021;11:796. [Crossref]
- Lavin A, Gilligan-Lee CM, Visnjic A, et al. Technology readiness levels for machine learning systems. Nat Commun 2022;13:6039. [Crossref] [PubMed]
- Clinical Decision Support Software: Guidance for Industry and Food and Drug Administration Staff. 2022:1-26. Available online: https://www.fda.gov/media/109618/download
- Administration USFaD. Your Clinical Decision Support Software: Is It a Medical Device? 2022. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/your-clinical-decision-support-software-it-medical-device
Cite this article as: Herrera Rivera N, McClintock DS, Alterman MA, Alterman TAL, Pruitt HD, Olsen GM, Harris NS, Marin MJ. A clinical laboratorian’s journey in developing a machine learning algorithm to assist in testing utilization and stewardship. J Lab Precis Med 2023;8:22.