
Reducing exposure to broad-spectrum antibiotics for bloodstream infection
The problem
Sepsis is an overwhelming, dysregulated inflammatory response caused by infection (1) and commonly results specifically from bloodstream infections (BSI). Annually, there are well over 19 million world-wide cases of sepsis with more than a million in the US (2). With annual costs exceeding $20 billion, sepsis is the most expensive condition treated in US hospitals (3). Timely treatment with antibiotics is so important that broad spectrum antibiotics are administered sometimes without knowledge of the source of infection and always without knowledge of the particular pathogen or its antibiotic susceptibility. According to the Surviving Sepsis Campaign Guidelines (4), empiric antimicrobial drugs with activity against all likely pathogens should be administered in the first hour of recognition of sepsis or septic shock. This shotgun approach is used because the current standard of care for pathogen detection, identification (ID) and antibiotic susceptibility (i.e., body fluid culture) is a very poor diagnostic. The time to result for blood cultures can exceed 48 hours. In addition, cultures have low sensitivity. When bacteria in the blood are in low numbers, growth is sufficiently slow to produce a negative result. In addition, certain bacteria do not grow at all under standard culture conditions (5). Worse still, cultures lack specificity and are routinely contaminated with normal flora of the skin (6). The result is a one-size-fits-all use of antibacterial drugs which leads to antibiotic resistance, opportunistic infection (e.g., Clostridium difficile), severe side effects (e.g., renal or hepatic failure) or potential under treatment of critically ill patients. A rapid diagnostic for pathogen ID and more importantly, antimicrobial susceptibility would reduce exposure time to empiric, broad-spectrum antibiotics and allow for rapid de-escalation to pathogen targeted therapy.
A unique yet critical subset of BSI worth noting are those associated with implanted medical devices. The cardiovascular devices market, specifically those with direct contact with the bloodstream (e.g., pacemakers, defibrillators, prosthetic heart valves, left ventricular assist devices, stents, vascular access devices, and venous filters), is one of the fastest growing medical device industries. This growth is a function of increasing prevalence of cardiovascular disease (7), expanding indications for implantation (8), and fundamental bioengineering improvements in materials and electronics (9). As indications for implantation continue to expand, patients with increasing comorbidities and decreasing immunocompetence will receive these devices. Despite the significant benefits afforded by these devices, any foreign material placed within the bloodstream poses a significant threat of infection. The underlying pathology is related to the adaptive strategy of bacteria to adhere to, colonize, and form biofilms on abiotic materials (10). These biofilms confer significant protection from both host immune clearance as well as antibiotics. While medical device implantation rates have increased by as much as 65% over the last few decades (11), infection rates have increased disproportionately (i.e., >200%) (12). As a result, implanted medical devices are a leading cause of BSI in the US. Novel antimicrobial/antibiofilm device strategies are being investigated, but the time horizon for their development, regulatory approval and clinical adoption is quite long. Therefore, the trend of increasing medical device associated BSI will likely continue or worsen in the near future.
A solution
With the recent adoption of the Sepsis Core Measure (SEP-1) by the Center for Medicare & Medicaid Service (CMS) there are strong economic incentives to increase vigilance for diagnosing sepsis. That combined with the current Sepsis-3 definitions (1) and Surviving Sepsis Guidelines (4) will continue to drive a low threshold for initiating empiric, broad-spectrum antibiotics. However, since the current diagnostic criteria are neither sensitive nor specific, there will be continued inappropriate use of broad- spectrum antibiotics with all the associated adverse effects (not the least of which is the continued selective pressure for antibiotic resistant species). This highlights the obvious clinical need to diagnosis BSI earlier and with improved accuracy including pathogen ID and antibiotic susceptibility. To that end, in a recent issue of Clinical Chemistry, Andini et al. (13) describe a nucleic acid amplification assay for detection, pathogen ID, and antibiotic susceptibility testing (AST) that shortens the time to result (and potentially the time to targeted antibiotic therapy) to ~7 hours. This is a profound improvement over the current standard of care which requires 48 hours or more (Figure 1).
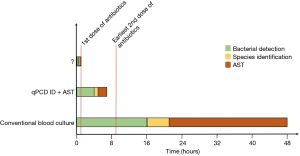
There are five key features of the described assay which contribute to it realized benefits. First, there is a custom initial sample processing step to remove human cells and DNA. This process is critical to remove inhibitors of polymerase chain reaction (PCR) as well as remove the large quantity of human DNA. In general, clinically significant bacteremia involves just a few bacterial cells per milliliter of blood. Therefore, there is a minuscule amount of bacterial DNA in a large background of human DNA. Removing the leukocyte DNA effectively improves the signal-to-noise ratio. Second, there is a brief culture enrichment period. Current quantitative PCR (qPCR) methods are not sufficiently sensitive to detect the very low concentrations of bacterial DNA directly from a blood sample. Therefore, the sample is cultured for 4 hours to allow bacteria to multiply to a level adequate for detection. Indeed, this allowed detection of 1 CFU/mL in the original sample. Third, the gene target for amplification by PCR was the 16–23 s ribosomal DNA (rDNA) internal transcribed spacer (ITS) region rather than the traditionally used 16 s rDNA. This sequence provides improved analytic specificity of different bacterial species. Fourth, the authors have developed a rich library of melt curves of the ITS region that effectively identify the bacterial species via high resolution melt curve analysis (HRMA) of the amplified gene product. In essence, this allows for sequential detection and ID without additional sample manipulation. Fifth, phenotypic AST that uses rapid qPCR to shorten the time to detect growth in the presence of antibiotics was incorporated. While many nucleic acid technologies focus on detection of antibiotic resistance genes, this technique evaluates the actual response of the bacteria to the antibiotic. This is more analogous to the currently accepted use of minimum inhibitory concentrations (MICs) and breakpoints for determining susceptibility. Furthermore, not all resistance mechanisms are known or related to single specific genetic sequences (e.g., efflux pumps) (14). Lastly, there is significant discordance between the presence of a particular resistance gene and phenotypic manifestation of resistance (15).
The value of appropriate vs. empiric antibiotics
Empiric antibiotics recommendations are based purely on covering the most likely pathogen without actual knowledge of the case specific pathogen. As such, it is prone to both over and under treatment of patients. Typically, if diagnostic testing is performed, the empiric antibiotic choice can be adjusted or de-escalated once the microbiologic testing is complete. Even shortening the time to result for such testing has significant benefits. For example, using the Verigene® technology from Nanosphere, Inc., Sango et al. demonstrated that reducing the time to detection of vancomycin-resistant enterococcus (VRE) by 23.4 hours lead to a 21.7-day reduction in length of stay and more than $60,000 reduction in health costs (16). This benefit was attributed to timely administration of appropriate antibiotics. However, that technology still depends on traditional blood cultures and at best had time to result of 18 hours, suggesting room for continued improvement. The benefit of appropriate pathogen-specific antibiotic therapy is further illustrated in a study comparing hospitalized patients with penicillin allergy versus non-allergic patients (17). It found that penicillin allergic patients received broader spectrum antibiotics such as fluoroquinolones, vancomycin, and clindamycin. They averaged 0.59 more total days in the hospital, had 23.4% more Clostridium difficile infections, 14.1% more methicillin resistant Staphylococcus aureus (MRSA) infections, and 30.1% more VRE infections. Over three years, the increased lengths of stay and complications cost the health care system ~$60 million. In general, maximal benefit of rapid molecular microbial testing is realized when paired with a well-designed antibiotic stewardship program. More specifically, the implementation of a rapid diagnostic bundle combined with evidence based antibiotic stewardship improved appropriateness of initial broad spectrum antibiotics and actually decreased broad-spectrum antibiotic utilization in a multi-hospital system (18). Overall, the use of rapid molecular diagnostics to improve time to appropriate, targeted therapy has shown positive clinical effects both in terms of patient outcomes and health care costs (19,20). Clearly, getting the right drug for the right bug has the potential to save lives and money.
The threat of emerging antibiotic resistance
The first antibiotic, penicillin, was discovered in 1928. Since then we have seen a tremendous improvement in global health and longevity. Unfortunately, emerging antimicrobial resistance is becoming one of the greatest threats to global human health. Less than two decades after his discovery of penicillin, Alexander Fleming noted that microbes become educated to resist penicillin (21). The time from development of a new class of antibiotics to the detection of resistance has been consistently decreasing (22). In a recent study of urinary tract infections diagnosed in the emergency department, up to 5% of positive cultures had extended-spectrum lactamase producing strains (23). Worse still, 46% of the patients had discordant initial empiric antibiotic coverage (i.e., antibiotics that do not cover the isolated strain). As more and more pathogenic bacteria develop resistance to multiple classes of antibiotics, previously treatable illnesses will become lethal. Addressing this challenge requires investment in not only new antimicrobial drug discovery but also rapid diagnostics and antibiotic stewardship to reduce the selective pressure for resistance (24).
Some words of caution
The combined qPCR-based ID + AST assay described by Andini et al. improves the time-to-result for ID and AST of a few BSI pathogens. This improvement surpasses both the current standard of care and many of the emerging technologies coming to market (14) in terms of both ID and phenotypic AST. As such, it is anticipated that, if implement on a larger scale, this assay could improve both individual patient outcomes as well as emerging antibiotic resistance by reducing the utilization of broad-spectrum antibiotics in favor of appropriate targeted therapy. That said, some caution is required with this optimism. First, the assay still relies on culture enrichment of the bacteria in Mueller-Hinton broth for detection. While dramatically reducing the culture time, the other limitations of blood culture (as noted above) remain. Specifically, like traditional blood cultures this system will have poor sensitivity for fastidious organisms and may under perform in biofilm infections where bacteria have reduced growth rates. Likewise, the potential for contamination by rapidly growing commensal skin strains remains high and therefore may lead to false positives. Second, the initial sample processing to remove leukocytes may reduce the potential detection of intracellular pathogens (e.g., Salmonella, Legionella, and Mycobacterium). Additionally, the exceedingly common BSI pathogen S. aureus, which was once considered an extracellular pathogen, can survive and even replicate within phagosomes thus evading immune detection and clearance (25). Therefore, under certain conditions, S. aureus may also evade detection by this strategy. Third, this proof-of-concept study only evaluated four common BSI pathogens. While this group represents roughly a third of all BSIs, the process for multiplex scale-up is certainly not trivial. Fourth, the direct inoculation method of AST used here represents a paradigm shift from conventional AST. Typically, bacterial growth in the presence of antibiotics is measured by optical density to determine susceptibility. However, this method is highly sensitive to the initial inoculum and therefore requires subculture from a positive blood culture bottle to obtain a standard concentration. The method described using a △Ct from qPCR to determine response to antibiotics removes the subculture step and is less sensitive to initial inoculum. While the use of direct inoculation methods is gaining traction there is significant inertia to challenge current standards to define resistance by MIC and breakpoints. The activation energy to replace this industry standard will be quite high. Taken together, these concerns suggest that the qPCR-based ID + AST system (though likely to be beneficial) will be unable to fully replace traditional blood culture. However, even as a purely additive strategy, rapid diagnostics coupled to antibiotic stewardship programs have demonstrate cost effectiveness (19).
A challenge for future work
At this point the significance of BSI including medical device associated BSI and resulting sepsis should be clearly evident. Furthermore, the potential benefits of improved diagnostics not just around predicting outcomes in sepsis but rather identifying culprit pathogens and their antibiotic susceptibility are equally obvious. In pursuing that goal, the assay described by Andini et al. is capable of reducing exposure to empiric broad spectrum antibiotics to a single dose (Figure 1). Indeed, it motivates a greater challenge. Can empiric broad spectrum antibiotics be eliminated all together? Will the continued improvement of diagnostic technologies allow for ID and AST in less than hour? Imagine a future where new antibiotics are narrow, even species specific and prescribed initially based on rapid, molecular, culture-free, diagnostics. The development of the qPCR ID + AST assay represents a collaboration between Emergency Medicine and Mechanical Engineering. This type of multidisciplinary approach is key to future success on this challenge. There are myriad of technologies at all developmental stages including chiroplasmonic nanomaterials (26,27), quantum tunneling enzymatic detection (28), magnetic resonance (29), and microfluidics (30) that may have impact in this space if given sufficient resources for multidisciplinary collaborative research and translation. Diagnostic-selected, narrow-spectrum antimicrobials are an obvious manifestation of precision medicine for infectious diseases and is clearly worthy of future pursuit and financial investment.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by our Section Editor Dr. Wei Li (Department of Clinical Laboratory, Qilu Hospital, Shandong University, Jinan, China).
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2018.12.02). Dr. VanEpps reports a patent, 16/761,686, COMPOSITIONS AND METHODS FOR DETECTION OF MICROORGANISMS, pending.The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- For sepsis, the drugs don’t work. Lancet Infect Dis 2012;12:89. [Crossref] [PubMed]
- Torio CM, Andrews RM. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2011. HCUP Statistical Brief #160. Available online: http://www.hcup-us.ahrq.gov/reports/statbriefs/sb160.pdf
- Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2016. Crit Care Med 2017;45:486-552. [Crossref] [PubMed]
- Peters RP, van Agtmael MA, Danner SA, et al. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 2004;4:751-60. [Crossref] [PubMed]
- Halverson S, Malani PN, Newton DW, et al. Impact of hourly emergency department patient volume on blood culture contamination and diagnostic yield. J Clin Microbiol 2013;51:1721-6. [Crossref] [PubMed]
- Mozaffarian D, Benjamin EJ, Go AS, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation 2016;133:e38-360. [PubMed]
- Praz F, Windecker S, Huber C, et al. Expanding Indications of Transcatheter Heart Valve Interventions. JACC Cardiovasc Interv 2015;8:1777-96. [Crossref] [PubMed]
- Khan W, Muntimadugu E, Jaffe M, et al. Implantable Medical Devices. In: Focal Controlled Drug Delivery. Domb AJ, Khan W, editors. Boston: Springer US;2014:33-59.
- VanEpps JS, Younger JG. Implantable Device-Related Infection. Shock 2016;46:597-608. [Crossref] [PubMed]
- Isaacs AJ, Shuhaiber J, Salemi A, et al. National trends in utilization and in-hospital outcomes of mechanical versus bioprosthetic aortic valve replacements. J Thorac Cardiovasc Surg 2015;149:1262-9.e3. [Crossref] [PubMed]
- Nielsen JC, Gerdes JC, Varma N. Infected cardiac-implantable electronic devices: prevention, diagnosis, and treatment. Eur Heart J 2015;36:2484-90. [Crossref] [PubMed]
- Andini N, Hu A, Zhou L, et al. A “Culture” Shift: Broad Bacterial Detection, Identification, and Antimicrobial Susceptibility Testing Directly from Whole Blood. Clin Chem 2018;64:1453-62. [Crossref] [PubMed]
- Opota O, Jaton K, Greub G. Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clin Microbiol Infect 2015;21:323-31. [Crossref] [PubMed]
- Owen JR, Noyes N, Young AE, et al. Whole-Genome Sequencing and Concordance Between Antimicrobial Susceptibility Genotypes and Phenotypes of Bacterial Isolates Associated with Bovine Respiratory Disease. G3 (Bethesda) 2017;7:3059-71. [Crossref] [PubMed]
- Sango A, McCarter YS, Johnson D, et al. Stewardship approach for optimizing antimicrobial therapy through use of a rapid microarray assay on blood cultures positive for Enterococcus species. J Clin Microbiol 2013;51:4008-11. [Crossref] [PubMed]
- Macy E, Contreras R. Health care use and serious infection prevalence associated with penicillin “allergy” in hospitalized patients: A cohort study. J Allergy Clin Immunol 2014;133:790-6. [Crossref] [PubMed]
- Bookstaver PB, Nimmich EB, Smith TJ 3rd, et al. Cumulative Effect of an Antimicrobial Stewardship and Rapid Diagnostic Testing Bundle on Early Streamlining of Antimicrobial Therapy in Gram-Negative Bloodstream Infections. Antimicrob Agents Chemother 2017;61. [PubMed]
- Pliakos EE, Andreatos N, Shehadeh F, et al. The Cost-Effectiveness of Rapid Diagnostic Testing for the Diagnosis of Bloodstream Infections with or without Antimicrobial Stewardship. Clin Microbiol Rev 2018;31. [PubMed]
- Buehler SS, Madison B, Snyder SR, et al. Effectiveness of Practices To Increase Timeliness of Providing Targeted Therapy for Inpatients with Bloodstream Infections: a Laboratory Medicine Best Practices Systematic Review and Meta-analysis. Clin Microbiol Rev 2016;29:59-103. [Crossref] [PubMed]
- Conly JM. Antimicrobial resistance - Judicious use is the key. Can J Infect Dis Med Microbiol 2004;15:249-51. [Crossref] [PubMed]
- Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T 2015;40:277-83. [PubMed]
- Frazee BW, Trivedi T, Montgomery M, et al. Emergency Department Urinary Tract Infections Caused by Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae: Many Patients Have No Identifiable Risk Factor and Discordant Empiric Therapy Is Common. Ann Emerg Med 2018;72:449-56. [Crossref] [PubMed]
- Perencevich EN, Malani PN. Treatment algorithms for staphylococcal bacteremia: Improving clinical care and enhancing antimicrobial stewardship. JAMA 2018;320:1243-4. [Crossref] [PubMed]
- Fraunholz M, Sinha B. Intracellular Staphylococcus aureus: live-in and let die. Front Cell Infect Microbiol 2012;2:43. [Crossref] [PubMed]
- Ma W, Kuang H, Xu L, et al. Attomolar DNA detection with chiral nanorod assemblies. Nat Commun 2013;4:2689. [Crossref] [PubMed]
- Zhao Y, Xu L, Ma W, et al. Shell-Engineered Chiroplasmonic Assemblies of Nanoparticles for Zeptomolar DNA Detection. Nano Letters 2014;14:3908-13. [Crossref] [PubMed]
- Shi X, Kadiyala U, VanEpps JS, et al. Culture-free bacterial detection and identification from blood with rapid, phenotypic, antibiotic susceptibility testing. Sci Rep 2018;8:3416. [Crossref] [PubMed]
- Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 2015;60:892-9. [Crossref] [PubMed]
- Hou HW, Bhattacharyya RP, Hung DT, et al. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 2015;15:2297-307. [Crossref] [PubMed]
Cite this article as: VanEpps JS. Reducing exposure to broad-spectrum antibiotics for bloodstream infection. J Lab Precis Med 2018;3:100.


