
miRNAs as novel biomarkers for bone related diseases
The history of microRNAs (miRNAs)
miRNAs contribute to the understanding of changes in bone metabolism. After their discovery as a new class of regulatory molecules they have become a focus of scientific interest (1). MiRNAs were first described in 1993 by Lee et al. (2). The first miRNAs identified were lin-4 RNA and let-7 RNA, both in Caenorhabditis elegans, where they are part of a programme that determines the timing of larval development (2,3). Mature miRNAs are 19 to 24 nucleotide long RNA molecules that are involved in regulatory processes intracellularly, but were detected also in the circulation and other body fluids (4).
Regulatory functions are executed by the binding of the so-called “seed” region of miRNAs, the 6–8 nucleotide long sequence that is energetically favourable for the interaction with the target mRNA. For their regulatory capacity, miRNA binding to target mRNA needs no perfect base pairing; therefore one miRNA can bind to more targets and control the expression of several mRNAs. In that way miRNA can control several mRNAs, but also several miRNAs can potentially target the same gene product. MiRNAs regulate gene expression by their complementary binding to the 3’ UTR (untranslated region) of the target RNA. Thereby, mRNA translation is hindered, or miRNA binding destabilizes the target mRNA, which is then cleaved and degraded (5,6).
Studies showed that miRNAs predominantly regulate gene expression negatively, at least in humans. Still, a discrepancy remains between the theoretically predicted binding sites and the actual binding of miRNAs leading to a biological response. Therefore, it has been suggested that the majority of predicted binding sites might not be relevant for biological processes (7,8).
The nomenclature of miRNAs
For the nomenclature of miRNAs a prefix indicates the organism, e.g., “hsa” for Homo sapiens, “mus” for Mus musculus etc. followed by the term “miR” and the identification number that is given according to the order of discovery, considering miRNAs with the same sequence carry the same number even in different organisms. In the case two miRNAs in the same organism share very similar, but not perfectly identical sequences, they can carry the same number, only with the addition of a small letter starting with “a” (9). Until today, the miRNA-database miRBase has 38,589 entries for 286 organisms. Mature miRNAs are listed as well as their precursors. It is suspected that these miRNAs regulate the translation of more than 60% of protein coding genes (9). miRNAs are, similar to transcription factors and RNA binding proteins, expressed tissue specific or at dedicated time points during development. This indicates that miRNAs are important factors in the development (10) and the cell-specific expression of protein profiles (11).
miRNA biogenesis
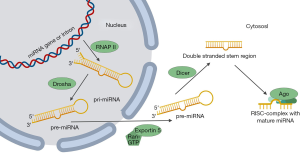
miRNAs either originate from non-protein coding transcripts (50%) or their sequence is located in the introns of coding genes (12). MiRNA biogenesis (Figure 1) starts in the nucleus with the transcription of a precursor miRNA (pri-miRNA) through RNA-polymerase II (RNAP II). The pri-miRNA has a length of a few thousand nucleotides and assembles to one or more hairpin elements. Via several ripening steps the pri-miRNA is processed into mature miRNA. The first maturation step is catalyzed by the nucleus located protein Drosha that cuts the stem-loop hairpin structure from the pri-miRNA yielding an approximately 70 nucleotide long hairpin structure called pre-miRNA (12). The protein exportin five complexes with the hairpin and together with a Ran-GTP (Ras-related nuclear protein-guanosine triphosphate) the pre-miRNA is actively released from the nucleus with the release of Ran-GDP and phosphate. Once in the cytosol, the second enzyme of the RNase-II family, Dicer, binds to the pre-miRNA and cuts off the loop of the hairpin, leaving the stem structure and the 19 to 22 nucleotide long miRNA duplex (13). Argonaute (AGO)-proteins bind to the structure and one of the two strands, the so-called passenger strand, is removed and a single stranded RNA-molecule bound to AGO-proteins remains. This complex is called the RISC-complex (RNA-induced silencing complex) (14). The active RISC-complex can then bind to the 3’UTR of the target mRNA by complementary base pairing of nucleotide 2 to 8 in the 5’ region of the miRNA. An imperfect base pairing can also lead to a silencing effect. Binding of the RISC-complex to the mRNA causes the inhibition of the protein synthesis via three different ways. Either miRNA binding marks the mRNA for degradation, or the translation is repressed and mRNA is de-adenylated or thirdly, RISC-binding alters DNA methylation and thereby regulates transcription directly (14).
Osteoporosis—an underestimated risk
The prevalence of osteoporosis for individuals in industrialized countries like North America, Europe, Japan and Australia at the age of 50 and above, as estimated from published data for the hip and/or the spine, ranged from 9% to 38% for women and 1% to 8% for men (15). The risk to develop osteoporosis is proportional to age (16). Women after menopause are at highest risk, as the absence of estrogen is one key factor for osteoporosis development (17). Vice versa, also the lack of androgenic hormones causes osteoporosis in men (18). The exact mode of action of these sex hormones on bone cells is still under discussion, but it is known that both bone-forming osteoblasts and bone-resorbing osteoclasts have receptors for both estrogens and androgens. Interestingly, estrogens protect cortical bone mass, whereas androgens are necessary for the maintenance of trabecular bone mass in male mammals, but not necessary for the anabolic effect on cortical bone (19). The lack of vitamin D is considered as a major factor for the reduction of bone mass although study outcomes are conflicting (20). Besides genetic predisposition there are many other risk factors for the development of osteoporosis such as metabolic diseases like diabetes mellitus type 1 and 2 (see review on diabetoporosity in this issue), anorexia nervosa, thyroid and renal dysfunctions or dietary as well as lifestyle habits such as low calcium intake or immobilization (21).
The most important outcomes of osteoporosis are low impact fractures, of which the distal forearm is the most common site of a first fracture in Caucasian women (22). Whereas vertebral fractures are frequently underdiagnosed, hip fractures can cause significant complications and mortality (23). A major problem is still the missing awareness and the late diagnosis of osteoporosis, leading into a downward spiral through consecutive fractures, loss of quality of life and even premature death of the patient (24). Early detection and rise of awareness could prevent fragility fractures (25) and a widespread screening of patients at risk could even help to lower the costs caused by the disease (26). However, diagnostic tools such as bone density measurements, bone turnover markers or fracture algorithms like FRAX have not been able to cover all the needs for an individual fracture risk prediction. Therefore, the availability of easy-to-measure biomarkers for osteoporosis, such as microRNAs (miRNAs) are considered to be an additional tool for the assessment of primary, as well as of secondary osteoporosis risk.
miRNAs as novel biomarkers
In 2008, two independent research teams identified the presence of miRNAs in the bloodstream (27,28). Since then, many different sequences have been found in human and animal derived plasma and serum (27). To date, these circulating miRNAs have been associated to many diseases, leading to the conclusion that miRNAs are “fingerprints” for specific diseases. Since miRNAs have been successfully annotated to specific biological functions and diseases, they soon became a novel class of biomarkers and even potential targets for therapies in the future (29-31).
Circulating miRNAs
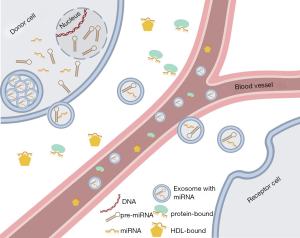
Chim et al. in 2008 were the first to describe circulating fetal nucleic acids in the bloodstream of the mother that were identified as miRNAs (32). Almost at the same time, tumour-associated miRNAs were detected in patients with diffuse B-cell lymphoma, which suggested for the first time their potential use as biomarkers (33). Circulating miRNAs can be found in several forms in the bloodstream (Figure 2): 90% of the extracellular miRNAs are bound to AGO-proteins (34,35) as free protein complexes in the bloodstream (34). The remaining 10% are packed in exosomes (36), in apoptotic bodies (35) or bound to HDL (high density lipoprotein) (37). The origin of circulating miRNAs is difficult to track. On the one hand, circulating miRNAs derive from dead blood cells, especially in the case of miRNAs bound to apoptotic bodies, but they can be also actively secreted by living cells (38). Interestingly, the stability of these circulating miRNAs is high because exosomes are impermeable for RNases and AGO-proteins are protecting the miRNAs from enzymatic disruption. Kept on −70 °C, miRNAs remain stable for at least one year (39,40). Since circulating miRNAs are stable for a certain time and easily accessible from body fluids, they are ideal candidates for their use as novel biomarkers.
Exosomes—miRNA carriers in bone disease
Exosomes are nanosized particles that are actively produced by a large number of cells and participate in a variety of signalling processes (41,42). Exosomes have been identified to transport numerous signalling molecules, such as proteins, signalling lipids and miRNAs (36,41). Exosomes have been associated to diverse physiological and pathophysiological conditions (42-45). Exosomes are produced from all types of bone cells and seem to play important roles in the differentiation process of osteoclasts and osteoblasts (46-48). In the recent years exosomes have been identified as important signal mediators in bone metabolism (48-50). Proteomic profiling from murine osteoblasts at various differentiation stages revealed a distinct proteomic signature at each time point, suggesting that distinct functions are mediated at each stage of osteoblast differentiation (51).
Some mechanistic studies were able to show an interplay of exosomal miRNAs and osteoblast or osteoclast function in bone degradation (52,53). Exosomal enriched miR-31a-5p was produced and released in high concentrations from bone marrow stromal cells (BMSC) of aged rats. It has been shown that miR-31a-5p expression in BMSCs increases with age and is actively transported to osteoclasts, increasing osteoclastogenesis and thereby contributes to age related bone loss. Antagonizing miR-31a-5p reversed the age-related phenotype and rescued bone loss (53). Similarly, a study by Sun et al. in 2016 showed that osteoclasts are able to inhibit osteoblastogenesis by secretion of exosomes carrying miR-214 in vitro and in vivo in mice. Inhibition of exosomes biogenesis in vitro diminished this effect. Moreover miR-214 abundance in serum of osteoporotic patients was increased compared to healthy controls and could serve as potential biomarker (52). Xie et al. characterized serum derived exosomes of elderly volunteers with osteoporosis or osteopenia. They found that the proteomic signatures of these serum derived exosomes favour osteoclast activation but also bone formation in osteopenia. The authors hypothesize that this might be a compensatory effect in osteopenia to counteract loss of bone mass. However, serum derived exosomes from patients with osteoporosis clearly induce osteoclast activation, inhibit osteoblast bone matrix mineralization and promote bone degradation (54).
Bone turnover is not only regulated via paracrine exosome delivery, also exosomes from distant tissue are able to affect bone formation (55-57). For instance exosomal miR-141-3p released from prostate cancer cells is able to control for osteoblast activity and induce bone metastasis (56). Qin et al. found that muscle secreted myostatin leads to suppression of miR-218 in osteocytes and released exosomes, which in turn reduced Runx2 expression in osteoblast and led to impaired osteoblastic differentiation (57).
Since exosomes and exosome associated miRNAs play pivotal roles in different aspects of bone turnover and associated pathologies, they represent promising and versatile tools for biomarker identification. Since exosomes transmit a variety of different signalling molecules from donor to recipient cells and act as so called “signalosomes”, they could be useful in identifying a group of biomarkers (41). Furthermore exosomes are found in almost all bodily fluids including blood, saliva, and urine and would allow for non-invasive liquid biopsy collection (58-60). Although many studies were able to identify exosomal cargo that is involved in bone formation and that might be able to serve as biomarker for bone loss, only a few studies exist that tried to identify exosomal markers in osteoporosis (52,53).
miRNAs as systemic biomarkers for osteoporosis
It is evident that miRNAs affect bone metabolism by influencing bone formation and resorption by targeting anabolic or catabolic processes. In the last few years, increasing evidence suggested miRNAs as biomarkers for osteoporosis, since they have been identified to play a crucial role in the interplay between osteoblasts and osteoclasts.
Circulating monocytes in vivo, a source of osteoclast precursors (61), showed an association of miR-133a with osteoporosis (62). MiR-133a and miR-21 were also suggested to be plasma biomarkers for osteoporosis (63). Whole blood lysates of postmenopausal Chinese women revealed several differentially regulated miRNAs, namely miR-130b-3p, miR-151a-3p, miR-151b, miR-194-5p, miR-590-5p, and miR-660-5p. Among them miR-194-5p was the most highly upregulated one with a more than 5-fold change. MiR-194-5p was identified to discriminate between osteoporosis and osteopenia (64), a precursor stage of osteoporosis (65). In another cohort of Chinese patients, circulating miR-125b, miR-30 and miR-5914 were upregulated and associated with postmenopausal osteoporosis (66). Another study investigated bone tissue of healthy and osteoporotic patients and found potential predictive miRNAs, namely miR-365, miR-10b, and miR-129-3p up-regulated and miRNA-671-5p, miR-141 and miR-25 down-regulated (67). A German research group showed that miR-21-5p, miR-93-5p, miR-100-5p, and miR-125b-5p were significantly upregulated in serum, tissue and bone cells of osteoporotic patients, independently of gender but directly correlated with BMD (68).
Among 790 miRNAs that could be detected 82 (~10%) were differentially expressed in a study that compared bone samples of freshly hip-fractured women with a control group of female osteoarthritic patients. Among the eight miRNAs with the lowest p-values two could be confirmed, miR-320a and miR-483-5p both targeting genes involved in bone metabolism (69).
A distinct miRNA-pattern in serum was found in patients with idiopathic osteoporosis. Eight miRNAs (miR-152-3p, miR-30e-5p, miR-140-5p, miR-324-3p, miR-19b-3p, miR-335-5p, miR-19a-3p, miR-550a-3p) showed an association to osteoporotic fractures regardless of age and sex of the study participants (70).
Osteoporosis is a disease that causes general bone loss and also the jaw and mandibles are affected by the disease. In ovariectomized mice, an established model for postmenopausal osteoporosis, mandibles and femurs were affected by the disease and a panel of miRNAs was differentially regulated in the mandible of the mice. The researchers suggested miR-17-5p and miR-133a-3p as the most promising biomarker candidates (71).
Of note, other non-coding RNA species are also able to add information on disease status apart from miRNAs, such as circular RNAs (circRNAs). One study found that hsa_circ_0001275 was negatively correlated with bone density T-scores and had significant diagnostic value in postmenopausal osteoporosis (72). Also, a species of long non-coding RNAs (lncRNAs) called DANCR was upregulated in female postmenopausal patients with low BMD (73).
Many studies show differentially expressed miRNAs in osteoporosis, however, this data suggest that not only one differentially regulated miRNA serves as biomarker. It rather may be different patterns of numerous miRNAs that can build an algorithm-based prediction. Furthermore, different research groups find quite different patterns, depending on the cohorts they compare. The retrospective approach of most studies limits the predictive power of the results. Furthermore, meta-analyses are still missing. In case of specific profiles and stable replication of the results, there are some promising attempts to use miRNA profiles for diagnostic purposes (Table 1).
Table 1
| miRNA(s) | Target(s) | Feature(s) |
|---|---|---|
| miRNAs and osteoblasts | ||
| miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-205, miR-217 | RUNX2 | Decrease of osteoblastic differentiation |
| miR-206 | CX43 | Overexpression inhibits osteoblast-differentiation |
| miR-29b | TGFβ, HDAC4, CTNNBIP1, DUSP | Promotes osteoblast differentiation by inhibiting neg. regulators |
| miR-433-3p | DKK1 | Promotes osteoblast differentiation |
| miR-145 | CBFB | Impairs bone regeneration |
| miR-103 | Runx2 | Increased by mechanotransduction |
| miRNAs and osteoclasts | ||
| miR-21 | PDCD4 | Promotes osteoclastogenesis |
| miR-223 | NFI-A | Promotes osteoclastogenesis |
| miR-146a | C-JUN, NF-ATc1, PU.1, TRAP | Impairs osteoclastogenesis |
| Disease-associated miRNAs in bone tissue | ||
| miR-2861 | HDAC5 | Promotes osteoblast differentiation, decreased levels in OPO-bones |
| miR-214 | ATF4 | Decreases osteoblast development |
| miR-21, miR-23a, miR-24, miR-25, miR-100, miR-125b | Overexpressed in serum and bone of recent hip fractured patients, affect osteogenic diff. of MSCs | |
| miR-22-3p, miR-328-3p, let-7g-5p | Different expression in serum of low impact hip fracture patients | |
| miRNAs as systemic biomarkers for osteoporosis | ||
| miR-21, miR-133a | Plasma conc. changed in OPO | |
| miR-130b-3p, miR-151a-3p, miR-151b, miR-194-5p, miR-590-5p, miR-660-5p | Plasma conc. changed in OPO | |
| MiR-194-5p | Differentiation of OPO and osteopenia | |
| miR-125b, miR-30, miR-5914 | Upregulated circulating levels associated with postmenopausal osteoporosis | |
| miR-365, miR-10b, miR-129-3p; miR-671-5p, miR-141, miR-25 | Bone tissue conc. upregulated in OPO; bone tissue conc. downregulated in OPO | |
| miR-21-5p, miR-93-5p, miR-100-5p, miR-125b-5p | Serum and bone conc. upregulated in OPO | |
| miR-320a, miR-483-5p | Changed in bone samples of acute hip-fracture | |
| miR-152-3p, miR-30e-5p, miR-140-5p, miR-324-3p, miR-19b-3p, miR-335-5p, miR-19a-3p, miR-550a-3p | Association in serum of acute hip fractures | |
| miR-17-5p, miR-133a-3p | Change in mouse mandibles of OVX-mice |
RUNX2, Runt-related transcription factor 2; CX43, connexin 43; TGF-β, transforming growth factor-beta; HDAC, histone deacetylase; CTNNBIP1, beta-catenin-interacting protein 1; DUSP, dual-specificity phosphatase; DKK1, dickkopf-related protein 1; CBFβ, core-binding factor subunit beta; PDCD4, programmed cell death protein 4; NFI-A, nuclear factor 1 A-type; NF-ATc1, nuclear factor of activated T-cells, cytoplasmic 1; TRAP, tartrate-resistant acid phosphatase; ATF4, activating transcription factor 4; OPO, osteoporosis; OVX, ovariectomy.
miRNA assay technologies
Several assays are available to perform miRNA profiling. The most common ones are summarized here in a short overview (74,75).
Quantitative polymerase chain reaction (qPCR) is the most flexible technique for small scale and versatile approaches. It is broadly used among different laboratories with established protocols and has high sensitivity and specificity. Therefore, qPCR is considered the technique for validation of other methods (75). In a first step, miRNAs are extracted from the biomaterial, ideally with protocols that enrich small RNA species to increase overall miRNA yield (76). With accurate primer selection, specificity can be highly increased. Primer design for miRNA qPCR is challenging, as mature miRNAs are only 22 nucleotides long, the same length than a traditional qPCR primer. Most solutions work by elongating the sequence for amplification. A specific primer for identifying a single miRNA is combined with a stem-loop/poly(A) reverse primer to elongate the PCR product. Both TaqMan® and SybrGreen® assays are available from various providers. Disadvantages of the technique are the labour-intensive workflow and the biased approach, as the miRNA has to be annotated for primer design (75). The selection of proper reference genes is still a challenge for miRNA qPCR. Although often addressed, the problem is not jet solved. Selection of classical reference genes like Act (Actin), ribosomal proteins or GAPDH (glyceraldehyde 3-phosphate dehydrogenase) are inappropriate in most cases as these mRNAs are way longer then miRNAs and behave differently in the extraction step. The use of short RNA species for normalization can also be problematic, as they might not be transcribed by the same polymerases as miRNA precursors. Theoretically, the best option would be a reference miRNA, but this miRNA is not defined jet. Sometimes, normalization candidates can be identified for a cohort after miRNA sequencing (77). If no reference genes are available, one solution for the normalization problem could be that no normalization is applied. In this case, extreme attention has to be paid on equivalent treatment of all the samples in one experiment. Spike-in miRNAs (synthetic miRNAs with known sequence) can be added for monitoring in each step of the preparation. Especially in experiments with more than 50 target genes per sample, the global mean method can be applied, where the mean miRNA content in each sample is used for normalization (74,78).
MiRNA sequencing is an unbiased approach, contrary to the biased approach via qPCRs. As such, this technique is an ideal tool for the discovery phase of an experiment. The system can distinguish between isoforms and closely related miRNAs. Although considered unbiased, the barcoding and pre-sequencing steps could add some bias to the experiment (79). It is the technique that requires the most input material and lacks the sensitivity of a qPCR approach. Also, data analysis can be quite challenging, especially when bioinformatics is not involved. Therefore, reproduction of sequencing results by qPCR is strongly recommended (80).
MiRNA arrays are widely used and available from different distributors. Even though they are not as quantitative compared to other techniques, they offer straight forward protocols and easy analyses. Hundreds of miRNAs can be tested at once at a reasonable price. Arrays are probed by hybridization of fluorescence labelled RNA or DNA samples. The brightness of individual spots corresponds to the expression and can be compared between samples. MiRNA arrays can be inaccurate if closely related sequences have to be distinguished. This can be counteracted partly by stringent washing and careful selection of the probes (81).
Multiplex miRNA profiling (e.g., FirePlexTM, NanoStringTM) is used increasingly in the recent years. The user-friendly technology offers the possibility to analyse multiple miRNAs in a variety of samples without extended protocols for other methods. Biofluids can be used directly, without pre-extraction of the miRNA species. Measurement of the assay is done by flow cytometers and data analysis is easier compared to sequencing (82,83).
miRNAs as regulators of bone metabolism
Bone turnover occurs continuously throughout life and is tightly balanced by bone building osteoblasts and bone resorbing osteoclasts. Recent studies highlighted the importance of miRNAs in both phases of bone turnover (84).
miRNAs and osteoblasts
Bone specific deletion of Dicer in a conditional mouse model causes fetal lethality and a deformed cartilaginous skeleton with no ossifications (85). Dicer or Drosha knockdown in human mesenchymal stem cells (MSCs) inhibited the differentiation to osteoblasts (86). Another study indicates, that a group of specific miRNAs, namely miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-205 and miR-217, lowered the expression of the osteogenic regulator Runx2 (Runt-related transcription factor 2) and therefore decreased osteoblastic differentiation (87). Negative effects on bone formation have also been shown for miR-206, which was found to be expressed in human osteoblasts. The levels of the miRNA decrease the further osteoblasts are differentiated. An overexpression of miR-206 inhibits osteoblast differentiation, whereas inhibition of miR-206 increases it. This effect is mediated by Cx43 (Connexin 43), a gap-junction protein in osteoblasts. Similarly in mice, overexpression of miR-206 in osteoblasts resulted in a lower bone mass and impaired osteoblast differentiation (88).
MiRNAs have been shown to induce osteoblast differentiation. MiR-29b inhibits negative regulators of the cellular signal cascade or of transcription programmes, including TGFβ (transforming growth factor beta) HDAC4 (histone-deacetylase 4) CTNNBIP1 (beta-catenin-interacting protein 1) and DUSP2 (dual specificity protein phosphatase 2). Therefore, miR-29b has a positive effect on the activity of signal cascades like Smad, ERK (extracellular signal-regulated kinase), MAPK (mitogen-activated protein kinase) or the Wnt signalling pathway. The activation of these pathways increases osteogenesis (89). miR-433-3p reduces the Dickkopf-1-protein, a potent antagonist in the canonical Wnt signalling. The expression of miR-433-3p is highly correlated to osteoblast differentiation and the expression of osteogenic markers Runx2 and osteocalcin in vitro (90). The binding partner of Runx2, Cbfb (core binding factor beta) is regulated by miR-145 and treatment of mouse femurs with miR-145 impaired bone regeneration after ablation of bone marrow (91).
A group of researchers was able to identify a connection between miRNAs and mechanotransduction. The authors demonstrated that miR-103 regulates Runx2, the master transcription factor of osteogenesis by responding to mechanical cues. Both, the miRNA and its host gene, PANK3 (pantothenate kinase 3) were upregulated during cyclic mechanical stretch (92) (Table 1).
miRNAs and osteoclasts
Opposed to miRNAs involved in osteoblast physiology, much less is known about the regulation of osteoclasts through miRNAs. However, some studies showed that knockdown of Dicer in mature murine osteoclasts caused increased bone mass in the trabecular regions and diminished the number of osteoclasts (93). This has been confirmed by another study, where mice with Dicer-knockdown in osteoclasts showed lower bone resorption with a decreased number of osteoclasts, but stable osteoblast number (94).
A feedback loop that positively influences osteoclast-maturation has been shown for miR-21. The transcription of this miRNA is induced by c-Fos, a transcription factor of the Fos-family. miR-21 inhibited PDCD4 (programmed cell death protein 4), leading to the activation of transcription factors that promote osteoclastogenesis, among them c-Fos, resulting in the initiation of macrophage differentiation to osteoclasts in the bone marrow (95).
In a cell model with osteoclast precursors, the transcription factor PU.1, induced by M-CSF (macrophage colony-stimulating factor) stimulated the production of RANK (receptor activator of NF-κB) and miR-223. miR-223 had a repressing effect on NFI-A (Nuclear factor 1 A-type), an inhibitor of M-CSFR (macrophage colony-stimulating factor receptor). This expression of M-CSFR is important for a physiological osteoclastogenesis and function (94).
MiR-146a negatively influenced the expression of several factors, among them c-Jun, NF-ATc1 (nuclear factor of activated T-cells, cytoplasmic 1), PU.1 and TRAP (tartrate-resistant acidic phosphatase), suggesting the conclusion that miR-146a has a negative effect on osteoclast development (96) (Table 1).
Disease-associated miRNAs in bone tissue
Soon after the discovery of miRNAs, this new group of regulatory molecules was associated with bone diseases. As soon as 2009, a study drew the connection between miR-2861 and osteoporosis. An overexpression of miR-2861 increased the BMP2 (bone morphogenetic protein 2) mediated osteoblastogenesis in mice. It has been shown that miR-2861 directly repressed HDAC5 (histone deacetylase 5). HDAC 5 in turn increased the degradation of Runx2. This suggested a positive influence of miR-2861 on osteoblast differentiation. A decreased miR-2861 expression is linked to a decreased rate of bone formation. Reduced levels of miR-2861 have been found in bone tissue of osteoporotic patients, too (97). Heterozygous missense mutations in WNT1, a key ligand in the Wnt-pathway, lead to a severe and early-onset form of osteoporosis. A pattern of circulating miRNAs could be linked to the mutation p.C218G in WNT1, suggesting a disrupted feedback regulation between the miRNAs and WNT1 (98).
An increased expression of miR-214 was associated with decreased bone formation in bone biopsies of fracture patients. Studies in mice confirmed that miR-214 influenced osteoblast differentiation and bone formation in bone diseases. miR-214 most likely affects ATF4 (activating transcription factor 4), a central transcription factor for the functional development of osteoblasts. Knock-down of miR-214 in contrast led to an increase in bone mineralization in mice (99). In a clinical study serum and bone tissue of patients with osteoporotic fractures was investigated regarding dysregulated miRNA expression. Five miRNAs were identified that were overexpressed in both bone and serum. miR-21, miR-23a, miR-24, miR-25, miR-100 and miR-125b (100). In a comparison to healthy controls patients with a recent low impact hip fracture showed significantly different expression levels of miR-22-3p, miR-328-3p and let-7g-5p in serum. This is particularly interesting as the same miRNAs are known to have an effect on bone metabolism in vitro (100). In vitro data support the correlation studies from Weilner et al., since all of the de-regulated miRNA species affected osteogenic differentiation of human MSCs.
Vascular calcification is a disease with similarity to bone formation and miRNAs play a critical role. Our group has shown that miRNAs related to bone are dysregulated during chronic kidney disease, a status that coincides with vascular calcification and demineralization of bones. Kidney transplantation has been shown to reverse this effects (101) (Table 1).
miRNAs in bone therapy
Since miRNAs harbour widespread functions in bone development and in the context of disease progression, miRNAs could be a possible target for novel therapies. Especially anti-miRs (miRNA inhibitors) and miRNA-mimics (synthetic miRNAs) are of interest in terms of their pharmacological and therapeutic use. Still, researchers have to carefully look at potential therapeutic applications of miRNAs, especially in terms of a systemic administration, because one miRNA can affect several targets and distinct tissues. As an example, miR-29a showed increased expression during osteoblast differentiation and regulates the differentiation process through the Wnt signalling pathway (102). MiR-29a protected bone formation from glucocorticoid-associated decrease in rats. Administration of miR-29a was able to prevent bone loss under glucocorticoid treatment (103). However, other studies identified miR-29a as an oncogenic factor that can be involved in tumour development. It increases the metastatic potential of colorectal cancer (104). A therapeutic use of miRNAs therefore has to be critically examined and the effect of the potential miRNA application or the knockdown of miRNAs with anti-miRs has to be checked by their characterization in different tissues.
miRNAs are key players in various aspects of bone metabolism. Several studies demonstrated the importance of miRNA regulation for osteoblast and osteoclast differentiation. Dysregulated miRNA expression in both cell types is involved in the development of bone disease such as osteoporosis. Since significant amounts of miRNAs (and other non-coding RNAs) can be found in the circulation, they provide unique potential for novel biomarkers: they show specificity for certain diseases and are easy accessible and stable molecules. Therefore, investigating miRNA- patterns in liquid biopsies represents a promising new avenue to early detection of osteoporosis and other bone diseases.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “Clinical and Analytical Aspects of Bone and Intersystemic Diseases”. The article has undergone external peer review.
Conflicts of Interest: The series “Clinical and Analytical Aspects of Bone and Intersystemic Diseases” was commissioned by the editorial office without any funding or sponsorship. Barbara Obermayer-Pietsch served as an unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Grabowski P. Physiology of Bone. Endocr Dev 2015;28:33-55. [Crossref] [PubMed]
- Lee RC, Feinbaum RL, Ambros V, et al. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000;403:901-6. [Crossref] [PubMed]
- Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 2010;11:537-61. [Crossref] [PubMed]
- Lewis BP, Shih I, Jones-Rhoades MW, et al. Prediction of Mammalian MicroRNA Targets. Cell 2003;115:787-98. [Crossref] [PubMed]
- Dweep H, Sticht C, Pandey P, et al. miRWalk - Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 2011;44:839-47. [Crossref] [PubMed]
- Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform 2014;15:1-19. [Crossref] [PubMed]
- Zhang L, Hammell M, Kudlow BA, et al. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development 2009;136:3043-55. [Crossref] [PubMed]
- Budak H, Bulut R, Kantar M, et al. MicroRNA nomenclature and the need for a revised naming prescription. Brief Funct Genomics 2016;15:65-71. [PubMed]
- Fernández-Pérez D, Brieño-Enríquez MA, Isoler-Alcaraz J, et al. MicroRNA dynamics at the onset of primordial germ and somatic cell sex differentiation during mouse embryonic gonad development. RNA 2018;24:287-303. [Crossref] [PubMed]
- Zhu W, Yang L, Du Z. MicroRNA regulation and tissue-specific protein interaction network. PLoS One 2011;6:e25394 [Crossref] [PubMed]
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415-9. [Crossref] [PubMed]
- Hutvágner G, Zamore PD. A microRNA in a Multiple-Turnover RNAi Enzyme Complex. Science 2002;297:2056-60. [Crossref] [PubMed]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22-32. [Crossref] [PubMed]
- Wade SW, Strader C, Fitzpatrick LA, et al. Estimating prevalence of osteoporosis: examples from industrialized countries. Arch Osteoporos 2014;9:182. [Crossref] [PubMed]
- Wright NC, Looker AC, Saag KG, et al. The Recent Prevalence of Osteoporosis and Low Bone Mass in the United States Based on Bone Mineral Density at the Femoral Neck or Lumbar Spine. J Bone Miner Res 2014;29:2520-6. [Crossref] [PubMed]
- Albright F, Smith PH, Richardson AM. Postmenopausal osteoporosis: Its clinical features. JAMA 1941;116:2465-74. [Crossref]
- Golds G, Houdek D, Arnason T. Male Hypogonadism and Osteoporosis: The Effects, Clinical Consequences, and Treatment of Testosterone Deficiency in Bone Health. Int J Endocrinol 2017;2017:4602129 [Crossref] [PubMed]
- Manolagas SC, O’Brien CA, Almeida M. The role of estrogen and androgen receptors in bone health and disease. Nat Rev Endocrinol 2013;9:699-712. [Crossref] [PubMed]
- Lips P, van Schoor NM. The effect of vitamin D on bone and osteoporosis. Best Pract Res Clin Endocrinol Metab 2011;25:585-91. [Crossref] [PubMed]
- Kanis JA, McCloskey EV. Risk factors in osteoporosis. Maturitas 1998;30:229-33. [Crossref] [PubMed]
- Poiana C, Carsote M, Radoi V, et al. Prevalent osteoporotic fractures in 622 obese and non- obese menopausal women. J Med Life 2015;8:462-6. [PubMed]
- Cannada LK, Hill BW. Osteoporotic Hip and Spine Fractures: A Current Review. Geriatr Orthop Surg Rehabil 2014;5:207-12. [Crossref] [PubMed]
- Hernlund E, Svedbom A, Ivergård M, et al. Osteoporosis in the European Union: medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos 2013;8:136. [Crossref] [PubMed]
- Harvey NCW, McCloskey EV, Mitchell PJ, et al. Mind the (treatment) gap: a global perspective on current and future strategies for prevention of fragility fractures. Osteoporos Int 2017;28:1507-29. [Crossref] [PubMed]
- Dimai HP, Redlich K, Peretz M, et al. Economic burden of osteoporotic fractures in Austria. Health Econ Rev 2012;2:12. [Crossref] [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [Crossref] [PubMed]
- Gilad S, Meiri E, Yogev Y, et al. Serum MicroRNAs Are Promising Novel Biomarkers. Williams S, editor. PLoS One 2008;3:e3148.
- Benz F, Roy S, Trautwein C, et al. Circulating MicroRNAs as Biomarkers for Sepsis. Int J Mol Sci 2016;17:78. [Crossref] [PubMed]
- Gupta SK, Bang C, Thum T. Circulating MicroRNAs as Biomarkers and Potential Paracrine Mediators of Cardiovascular Disease. Circ Cardiovasc Genet 2010;3:484-8. [Crossref] [PubMed]
- Dangwal S, Stratmann B, Bang C, et al. Impairment of Wound Healing in Patients With Type 2 Diabetes Mellitus Influences Circulating MicroRNA Patterns via Inflammatory Cytokines. Arterioscler Thromb Vasc Biol 2015;35:1480-8. [Crossref] [PubMed]
- Chim SSC, Shing TKF, Hung ECW, et al. Detection and Characterization of Placental MicroRNAs in Maternal Plasma. Clin Chem 2008;54:482-90. [Crossref] [PubMed]
- Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672-5. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci 2011;108:5003-8. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Sohel MH. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev Life Sci 2016;10:175-86. [Crossref]
- Sourvinou IS, Markou A, Lianidou ES. Quantification of Circulating miRNAs in Plasma. J Mol Diagn 2013;15:827-34. [Crossref] [PubMed]
- Li Y, Jiang Z, Xu L, et al. Stability analysis of liver cancer-related microRNAs. Acta Biochim Biophys Sin (Shanghai) 2011;43:69-78. [Crossref] [PubMed]
- Record M, Subra C, Silvente-Poirot S, et al. Exosomes as intercellular signalosomes and pharmacological effectors. Biochem Pharmacol 2011;81:1171-82. [Crossref] [PubMed]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. [Crossref] [PubMed]
- Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017;542:450-5. [Crossref] [PubMed]
- Martin-Medina A, Lehmann M, Burgy O, et al. Increased Extracellular Vesicles Mediate WNT-5A Signaling in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Becker A, Thakur BK, Weiss JM, et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016;30:836-48. [Crossref] [PubMed]
- Qin Y, Peng Y, Zhao W, Pan J, et al. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. J Biol Chem 2017;292:11021-33. [Crossref] [PubMed]
- Yuan FL, Wu QY, Miao ZN, et al. Osteoclast-Derived Extracellular Vesicles: Novel Regulators of Osteoclastogenesis and Osteoclast-Osteoblasts Communication in Bone Remodeling. Front Physiol 2018;9:628. [Crossref] [PubMed]
- Behera J, Tyagi N. Exosomes: mediators of bone diseases, protection, and therapeutics potential. Oncoscience 2018;5:181-95. [PubMed]
- Cui Y, Luan J, Li H, et al. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. Chang Z, editor. FEBS Lett 2016;590:185-92.
- Holliday LS, McHugh KP, Zuo J, et al. Exosomes: novel regulators of bone remodelling and potential therapeutic agents for orthodontics. Orthod Craniofac Res 2017;20:95-9. [Crossref] [PubMed]
- Bilen MA, Pan T, Lee YC, et al. Proteomics Profiling of Exosomes from Primary Mouse Osteoblasts under Proliferation versus Mineralization Conditions and Characterization of Their Uptake into Prostate Cancer Cells. J Proteome Res 2017;16:2709-28. [Crossref] [PubMed]
- Sun W, Zhao C, Li Y, et al. Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Cell Discov 2016;2:16015. [Crossref] [PubMed]
- Xu R, Shen X, Si Y, et al. MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment. Aging Cell 2018;17:e12794 [Crossref] [PubMed]
- Xie Y, Gao Y, Zhang L, et al. Involvement of serum-derived exosomes of elderly patients with bone loss in failure of bone remodeling via alteration of exosomal bone-related proteins. Aging Cell 2018;17:e12758 [Crossref] [PubMed]
- Tofiño-Vian M, Guillén MI, Pérez Del Caz MD, et al. Extracellular Vesicles from Adipose-Derived Mesenchymal Stem Cells Downregulate Senescence Features in Osteoarthritic Osteoblasts. Oxid Med Cell Longev 2017;2017:7197598 [Crossref] [PubMed]
- Ye Y, Li SL, Ma YY, et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 2017;8:94834-9. [PubMed]
- Qin Y, Peng Y, Zhao W, et al. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. J Biol Chem 2017;292:11021-33. [Crossref] [PubMed]
- Li X, Seebacher NA, Hornicek FJ, et al. Application of liquid biopsy in bone and soft tissue sarcomas: Present and future. Cancer Lett 2018;439:66-77. [Crossref] [PubMed]
- Michael A, Bajracharya SD, Yuen PST, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis 2010;16:34-8. [Crossref] [PubMed]
- Huebner AR, Somparn P, Benjachat T, et al. Exosomes in Urine Biomarker Discovery. Adv Exp Med Biol 2015;845:43-58. [Crossref] [PubMed]
- Zhou Y, Deng HW, Shen H. Circulating monocytes: an appropriate model for bone-related study. Osteoporos Int 2015;26:2561-72. [Crossref] [PubMed]
- Wang Y, Li L, Moore BT, et al. MiR-133a in Human Circulating Monocytes: A Potential Biomarker Associated with Postmenopausal Osteoporosis. PLoS One 2012;7:e34641 [Crossref] [PubMed]
- Li H, Wang Z, Fu Q, et al. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers 2014;19:553-6. [Crossref] [PubMed]
- Meng J, Zhang D, Pan N, et al. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. PeerJ 2015;3:e971 [Crossref] [PubMed]
- Kanis JA, Johnell O, Oden A, et al. Risk of hip fracture according to the World Health Organization criteria for osteopenia and osteoporosis. Bone 2000;27:585-90. [Crossref] [PubMed]
- Chen H, Jiang H, Can D, et al. Evaluation of MicroRNA 125b as a potential biomarker for postmenopausal osteoporosis. Trop J Pharm Res 2017;16:641. [Crossref]
- Zhu Y, Teng Z, Liu Y, et al. Six miRNAs identified serving as prognostic and predictive markers for osteoporosis by miRNA high-throughput method. Int J Clin Exp Med 2016;9:15226-34.
- Kelch S, Balmayor ER, Seeliger C, et al. MiRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci Rep 2017;7:15861. [Crossref] [PubMed]
- De-Ugarte L, Yoskovitz G, Balcells S, et al. MiRNA profiling of whole trabecular bone: Identification of osteoporosis-related changes in MiRNAs in human hip bones. BMC Med Genomics 2015;8:75. [Crossref] [PubMed]
- Kocijan R, Muschitz C, Geiger E, et al. Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab 2016;101:4125-34. [Crossref] [PubMed]
- Hao L, Li J, Tian Y, et al. Changes in the MicroRNA Profile of the Mandible of Ovariectomized Mice. Cell Physiol Biochem 2016;38:1267-87. [Crossref] [PubMed]
- Zhao K, Zhao Q, Guo Z, et al. Hsa_Circ_0001275: A Potential Novel Diagnostic Biomarker for Postmenopausal Osteoporosis. Cell Physiol Biochem 2018;46:2508-16. [Crossref] [PubMed]
- Tong X, Gu P, Xu S, et al. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem 2015;79:732-7. [Crossref] [PubMed]
- Baker M. MicroRNA profiling: separating signal from noise. Nat Methods 2010;7:687-92. [Crossref] [PubMed]
- van Rooij E. The art of MicroRNA research. Circ Res 2011;108:219-34. [Crossref] [PubMed]
- McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 2011;57:833-40. [Crossref] [PubMed]
- Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA 2012;3:601-16. [Crossref] [PubMed]
- Bockmeyer CL, Säuberlich K, Wittig J, et al. Comparison of different normalization strategies for the analysis of glomerular microRNAs in IgA nephropathy. Sci Rep 2016;6:31992. [Crossref] [PubMed]
- Alon S, Vigneault F, Eminaga S, et al. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res 2011;21:1506-11. [Crossref] [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: Approaches and considerations. Nat Rev Genet 2012;13:358-69. [Crossref] [PubMed]
- Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem 2009;394:1117-24. [Crossref] [PubMed]
- Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 2014;11:809-15. [Crossref] [PubMed]
- Tackett MR, Diwan I. Using FirePlexTM Particle Technology for Multiplex MicroRNA Profiling Without RNA Purification. New York, NY: Humana Press, 2017:209-19.
- Valenti MT, Dalle Carbonare L, Mottes M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease Int J Mol Med 2018;41:2441-9. (Review). [PubMed]
- Gaur T, Hussain S, Mudhasani R, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol 2010;340:10-21. [Crossref] [PubMed]
- Oskowitz AZ, Lu J, Penfornis P, et al. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci U S A 2008;105:18372-7. [Crossref] [PubMed]
- Zhang Y, Xie RL, Gordon J, et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem 2012;287:21926-35. [Crossref] [PubMed]
- Inose H, Ochi H, Kimura A, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci 2009;106:20794-9. [Crossref] [PubMed]
- Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem 2009;284:15676-84. [Crossref] [PubMed]
- Tang X, Lin J, Wang G, et al. MicroRNA-433-3p promotes osteoblast differentiation through targeting DKK1 expression. PLoS One 2017;12:e0179860 [Crossref] [PubMed]
- Fukuda T, Ochi H, Sunamura S, et al. MicroRNA-145 regulates osteoblastic differentiation by targeting the transcription factor Cbfb. FEBS Lett 2015;589:3302-8. [Crossref] [PubMed]
- Zuo B, Zhu JF, Li J, et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J Bone Miner Res 2015;30:330-45. [Crossref] [PubMed]
- Mizoguchi F, Izu Y, Hayata T, et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem 2010;109:866-75. [PubMed]
- Sugatani T, Hruska KA. Impaired Micro-RNA Pathways Diminish Osteoclast Differentiation and Function. J Biol Chem 2009;284:4667-78. [Crossref] [PubMed]
- Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood 2011;117:3648-57. [Crossref] [PubMed]
- Nakasa T, Shibuya H, Nagata Y, et al. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum 2011;63:1582-90. [Crossref] [PubMed]
- Li H, Xie H, Liu W, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 2009;119:3666-77. [Crossref] [PubMed]
- Mäkitie RE, Hackl M, Niinimäki R, et al. Altered MicroRNA Profile in Osteoporosis Caused by Impaired WNT Signaling. J Clin Endocrinol Metab 2018;103:1985-96. [Crossref] [PubMed]
- Wang X, Guo B, Li Q, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med 2013;19:93-100. [Crossref] [PubMed]
- Weilner S, Skalicky S, Salzer B, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone 2015;79:43-51. [Crossref] [PubMed]
- Ulbing M, Kirsch AH, Leber B, et al. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone 2017;95:115-23. [Crossref] [PubMed]
- Kapinas K, Kessler C, Ricks T, et al. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 2010;285:25221-31. [Crossref] [PubMed]
- Wang FS, Chung PC, Lin CL, et al. MicroRNA-29a Protects Against Glucocorticoid-Induced Bone Loss and Fragility in Rats by Orchestrating Bone Acquisition and Resorption. Arthritis Rheum 2013;65:1530-40. [Crossref] [PubMed]
- Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer 2014;110:450-8. [Crossref] [PubMed]
Cite this article as: Foessl I, Kotzbeck P, Obermayer-Pietsch B. miRNAs as novel biomarkers for bone related diseases. J Lab Precis Med 2019;4:2.


