The current and future role of laboratory medicine for diagnosing gestational diabetes mellitus
Introduction
Gestational diabetes mellitus (GDM) is a complication of pregnancy, defined as the “diabetes diagnosed in the second or third trimester of pregnancy that was not clearly overt diabetes prior to gestation” (1). The prevalence of this condition approximates 14% of worldwide pregnancies, but varies widely from 1% to 28% depending on population characteristics (e.g., maternal age, socioeconomic status, race/ethnicity and body composition) and diagnostic criteria (2-4). Caucasian women are at lower risk of developing GDM compared to Native Americans, Hispanics, Asians, and African-American women (5), though the prevalence significantly increases in women with predisposing conditions or risk factors. In a large European multicenter study based on a cohort of women with body mass index (BMI) ≥29 kg/m2, the prevalence of GDM was found to approximate 40% using the International Association of Diabetes and Pregnancy Study Groups (IADPSG)/World Health Organization (WHO) 2013 diagnostic criteria (6).
The pathophysiology of GDM is characterized by progressive development of insulin resistance, mainly triggered by placental production of diabetogenic hormones such as estrogen, progesterone, leptin, cortisol, placental lactogen and placental growth hormone (7). GDM develops in predisposed pregnant women in whom the adaptation β-cell hyper-functionality fails to compensate maternal insulin resistance (8). Risk factors for GDM include classical risk factors for metabolic as well as for cardiovascular diseases (CVDs) (i.e., overweight/obesity, advanced age, gestational weight gain, westernized diet, a family history of insulin resistance and/or diabetes) (9,10), but also other non-typical risk factors, such as ABO blood group (11) or genetic (12,13) and autoimmune conditions (14).
Although GDM typically resolves with and after delivery, early diagnosis and consequent management (i.e., lifestyle changes followed by oral blood-glucose-lowering agents or insulin, if necessary) are essential for lowering the maternal risk of developing type 2 diabetes mellitus (T2DM) (15), fatty liver disease (16), metabolic syndrome (17) and CVD (18), along with other short-term and long-term complications for the offspring (i.e., high birth weight, congenital malformations, intrauterine growth restriction and preterm birth, respiratory distress syndrome, and so on) (19,20). In keeping with this evidence, the Hyperglycemia and Adverse Pregnancy Outcome Follow-up Study (HAPO FUS) has recently confirmed that GDM is independently associated with childhood impaired glucose tolerance (IGT) (21).
The current screening/diagnostic approach
Several guidelines have been developed during the past decades, which differ for timing of oral glucose tolerance test (OGTT), population tested, number of samples analyzed, glucose load and glucose thresholds (22) (Table 1). Although universal agreement has not been reached, the most followed guidelines are those recommending one-step 75 g OGTT strategy, following the IADPSG criteria (i.e., to be performed between 24–28 gestational weeks) (36). This approach has been proposed after the publication of the HAPO study, including more than 23,000 pregnant women, which demonstrate that that complications for mother and offspring increases in parallel with maternal glycaemia at 24–28 weeks of gestation (37). According to IADPSG criteria, one value exceeding the following established cutoffs is sufficient for diagnosing GDM: 5.1 mmol/L (92 mg/dL) for fasting plasma glucose, 10 mmol/L (180 mg/dL) after 1 hour and 8.5 mmol/L (153 mg/dL) after 2 hours from the glucose intake (38). Since it has been demonstrated that selective screening based on traditional risk factors for GDM has a relatively low sensitivity (39), the screening approach has been extended to the entire population of pregnant women not only by the American Diabetes Association (ADA), but also by other international scientific organizations such as the WHO, the International Diabetes Federation (IDF) and the International Federation of Gynecology and Obstetrics (FIGO).
Table 1
| Year | References | Number of steps | Oral glucose load | Number of abnormal values required | Glucose cut-offs |
|---|---|---|---|---|---|
| 1964 | O’Sullivan et al. (23) | 1 | 100 g | ≥2 | Fasting ≥2 SD above the mean |
| 1 h ≥2 SD above the mean | |||||
| 2 h ≥2 SD above the mean | |||||
| 3 h ≥2 SD above the mean | |||||
| 1973 | O’Sullivan et al. (24) | 2 | First-step: 50 g; second-step: 100 g | ≥2 | Fasting ≥5.0 mmol/L (90 mg/dL) |
| 1 h ≥9.2 mmol/L (165 mg/dL) | |||||
| 2 h ≥8.1 mmol/L (145 mg/dL) | |||||
| 3 h ≥7.0 mmol/L (125 mg/dL) | |||||
| 1979 | NDDG (25) | 1 | 100 g | ≥2 | Fasting ≥5.9 mmol/L (105 mg/dL) |
| 1 h ≥10.6 mmol/L (190 mg/dL) | |||||
| 2 h ≥9.2 mmol/L (165 mg/dL) | |||||
| 3 h ≥8.1 mmol/L (145 mg/dL) | |||||
| 1982 | Carpenter et al. (26) | 2 | First-step: 50 g; second-step: 75 g | ≥2 | Fasting ≥5.3 mmol/L (95 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.6 mmol/L (155 mg/dL) | |||||
| 1996 | EASD (27) | 1 | 75 g | ≥1 | Fasting ≥6.0 mmol/L (108 mg/dL) |
| 2 h ≥9.0 mmol/L (162 mg/dL) | |||||
| 1999 | WHO (28) | 1 | 75 g | ≥1 | Fasting ≥7.0 mmol/L (126 mg/dL) |
| 2 h ≥7.8 mmol/L (140 mg/dL) | |||||
| 2007 | Metzger et al. (29) | 1 | 100 g | ≥2 | Fasting ≥5.3 mmol/L (95 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.6 mmol/L (155 mg/dL) | |||||
| 3 h ≥7.8 mmol/L (140 mg/dL) | |||||
| 2000 | ADA (30) | 1 | 75/100 g | ≥2 | Fasting ≥5.3 mmol/L (95 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.6 mmol/L (155 mg/dL) | |||||
| 3 h ≥7.8 mmol/L (140 mg/dL) | |||||
| 2010 | IADPSG and ADA (31) | 1 | 75 g | ≥1 | Fasting ≥5.1 mmol/L (92 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.5 mmol/L (153 mg/dL) | |||||
| 2013 | WHO (32) | 1 | 75 g | ≥1 | Fasting ≥5.1 mmol/L (92 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.5 mmol/L (153 mg/dL) | |||||
| 2015 | NICE (33) | 1 | 75 g | ≥1 | Fasting ≥5.6 mmol/L (100 mg/dL) |
| 2 h ≥7.8 mmol/L (140 mg/dL) | |||||
| 2015 | FIGO (34) | 1 | 75 g | ≥1 | Fasting ≥5.1 mmol/L (92 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.5 mmol/L (153 mg/dL) | |||||
| 2018 | ACOG (35) | 2 | First-step: 50 g; second-step: 100 g | ≥2 | Fasting ≥5.3 mmol/L (95 mg/dL) |
| 1 h ≥10.0 mmol/L (180 mg/dL) | |||||
| 2 h ≥8.6 mmol/L (155 mg/dL) | |||||
| 3 h ≥7.8 mmol/L (140 mg/dL) |
ACOG, American College of Obstetricians and Gynecologists; GDM, gestational diabetes mellitus; IADPSG, International Association of Diabetes in Pregnancy Study Groups; NICE, National Institute for Health and Care Excellence; OGTT, oral glucose tolerance test; WHO, World Health Organization; EASD, European Association for the Study of Diabetes; NDDG, National Diabetes Data Group; ADA, American Diabetes Association; FIGO, International Federation of Gynecology and Obstetrics; SD, standard deviation.
Unlike other countries which actually follow the recommendations of ADA, WHO, IDF and FIGO, the Italian National Health System guidelines in 2011 has limited the screening to women at risk of GDM rather than endorsing an approach based on universal screening (40). According to this Italian guideline, high risk women (i.e., previous GDM, pre-pregnancy BMI ≥30 kg/m2, FPG 100–125 mg/dL in the first trimester of pregnancy) should be screened between the 16th and 18th gestational weeks, which screening repeated between the 24th and 28th gestational weeks in the presence of normal glucose tolerance, whilst in women with medium risk (i.e., age ≥35 years, pre-pregnancy BMI 25–29.9 kg/m2, family history of T2DM, previous macrosomia and of an ethnic group at GDM risk) screening is only recommended between the 24th and 28th gestational weeks (40). Recently published studies showed that this approach is characterized by low sensitivity for detecting GDM, thus emphasizing the real need of a substantially critical revision (41,42).
Another aspect that merits consideration is the preanalytical quality of the samples, which include reliable conditions of storage, sample management and use of suitable glycolysis inhibitors (43-46). Screening and diagnostic tests can be performed more or less accurately, thus potentially increasing the number of false negatives (47).
Epigenetics changes as potential biomarkers of GDM
Epigenetics plays a substantial role in the pathogenesis of several conditions, as well as in disorders of glucose metabolism (48-51). It has also been hypothesized that some epigenetic mechanisms, including DNA methylation, histone modifications and small non-coding RNAs, could fill the knowledge gap between environmental factors (i.e., diet, pollution, stress, smoke and others) and heritable genetic susceptibility (52).
DNA methylation is a reversible process consisting of addition of a methyl group to the fifth carbon position of a cytosine residue within cytosine-phosphate-guanine (CpG) dinucleotides (a process catalyzed by the enzyme DNA methyltransferase), thus inhibiting gene transcription (53). Both aberrant global methylation and DNA methylation of specific genes involved in insulin resistance, for example in response to nutritional and environmental factors, have been linked to the pathogenesis of GDM (54). On the other hand, it has been suggested that also GDM may have an impact on epigenetic modifications in mother and offspring (55).
Several studies investigating the role of epigenetics in the pathogenesis of GDM have been originally carried out in the animal model (56-59). More recently, these epigenetic modifications have been also demonstrated in different human tissues, including placenta, cord blood, as well as in visceral and subcutaneous adipose tissues. Placenta DNA methylation of more than 385,000 CpG sites has been assessed by Rong et al. (60) in 36 GDM women and 40 controls. The authors identified both hyper- and hypo-methylated regions in GDM patients compared to healthy subjects, thus hypothesizing a role of this epigenetic modification in the pathophysiology of GDM. The differentially methylated genes, IGF2, GCKR and KCNQ1, are involved in pathways of cell growth, death regulation, immune and inflammatory response and nervous system development (60).
Although Nomura et al. failed to find an association between methylation status and GDM in an earlier study based on 50 placenta samples (61), Reichetzeder et al. (62) analyzed a larger number of placental tissues (n=1,030) by means of liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), observing that placental global DNA hypermethylation was associated with GDM, independently from other risk factors. Deng et al. (63) conducted a global gene methylation and whole genome expression profiling in visceral omental adipose tissue of GDM and normal pregnancies. The authors found that 935 genes were commonly dysregulated in the GDM group compared to healthy pregnant women (63).
Since it has been shown that maternal peripheral blood reflects placental epigenetics changes (64), several studies have been carried out for evaluating methylation status in blood. In a study performed in peripheral blood cells of 63 South African women with GDM, no difference in global DNA methylation could be observed between women with or without GDM (65). In this study the analysis was performed in women between the 24–28 gestational weeks, and it is hence conceivable that differences in methylation would be more clearly evident when measured earlier. In keeping with this hypothesis, Enquobahrie et al. (66) studied GDM women before the 20th gestational week, and reported that 17 CpG sites were hypomethylated, whilst 10 CpG sites were found to be hypermethylated. Even more interestingly, a recent study identified a characteristic genome-wide DNA methylation profiling measured between the 12th and 16th gestational weeks in 11 GDM women compared to 11 matched controls (67). In particular, five genes (COPS 8, PIK3R5, HAAO, CCDC124, and C5orf34) displayed a significantly different methylation status (67). Kang et al. carried out a genome-wide DNA methylation profile on 8 women with GDM and 8 healthy controls, observing as many as 151 genes with different degree of methylation (68). Among these, genes codifying for pro-inflammatory cytokine interleukin-6 (IL-6) and for anti-inflammatory cytokine IL-10 were identified. In addition to studying the global methylation profile, another approach involves the study analysis of methylation of single genes, though locus-specific DNA methylation methods are expensive and require large bioinformatics expertise (69). The gene codifying for leptin is among those most studied in GDM, since it is finely regulated by many epigenetic mechanisms (70). In two subsequent studies, Bouchard et al. (71,72) observed an association between hyperglycemia and alterations in placental DNA methylation of leptin and adiponectin genes in a cohort of mothers with impaired glucose metabolism. Kang et al. enrolled a Taiwanese population encompassing of 8 GDM and 24 controls, and found decreased methylation of IL-10 in blood of GDM, which was also found to be associated with increased serum values of IL-10 (73). More recently, Zhang et al. studied the methylation level of HIF3A promoter region in 20 GDM patients, showing that HIF3A expression is down-regulated in omental tissues (74). Notably, previous studies showed that HIF3A is involved in insulin resistance and glucose metabolism, thus providing a reasonable support to these findings (75).
Although translation of methylation study into clinical practice is challenging, microRNAs (miRNAs)—small non-coding RNAs (approximately 20 nucleotides in length) that regulate gene expression—can be seen as potentially useful circulating biomarkers for monitoring pregnancy and screening GDM. Studies performed using placenta tissues revealed that placenta has a specific miRNA expression pattern (76,77), and that this profile dynamically changes during pregnancy (78). A number of studies have explored differential expression of miRNAs at delivery in GDM pregnancies versus healthy controls to date (79-84), whilst other studies have investigated the potential clinical usefulness of miRNAs deregulation in assessing the risk of developing GDM though measurement of circulating miRNA levels in first or second-trimester in women with and without GDM (85-90).
In 2011, Zhao et al. (85) studied 24 GDM pregnant women (16–19 gestational week) and 24 healthy pregnant women, and identified three miRNAs (miR-132, miR-29a and miR-222) significantly down-regulated in GDM. In another study based on 28 women with GDM and 53 controls, down-regulation of miR-222, associated with low expression of miR-20a, was also confirmed in the study of Pheiffer and colleagues (89). Unlike these findings, Wander et al. (88) failed to find significant differences in plasma levels of miR-222 and miR-29a assayed in 36 GDM cases and 80 controls from the Omega prospective study. More interestingly, Wander et al. reported enhanced circulating values of miR-155 and miR-21 in GDM women, especially in overweight/obese pre-pregnancy or pregnant with male offspring (88). MiR-222 expression was also studied by Shi et al. (84) in omental adipose tissue of GDM patients and was found to be over-expressed. Moreover, miR-222 levels were positively correlated with maternal estradiol concentrations and negatively with estrogen receptor, thus reinforcing the idea of a role of this miR in the pathogenesis of insulin resistance.
These findings were confirmed in the recent study of Tagoma et al. (90), who found that miR-222 and several other miRNAs were over-expressed in maternal plasma of women with GDM. Other case-controls studies, mainly based on small sized populations (86,87,91-94) identified a number of other miRNAs that were deregulated in pregnant women with GDM.
Despite many studies could identify many miRNAs which can be potentially used as diagnostic biomarkers, the real utility of this approach in clinical practice has not been demonstrated so far, and larger well-performed studies are needed. Moreover, consensus is lacking on several pre-analytical and analytical aspects (i.e., biological matrix, sample handing procedure, sample storage, quantification technique, data normalization, etc.), which finally preclude results comparability. Important standardization or harmonization efforts shall hence be planned to overcome these current drawbacks (95,96).
Future perspectives
A new class of endogenous ncRNA biomarkers, named circular RNAs (circRNAs), has been investigated in different diseases during the past decade, including age-related pathologies such as cancer, CV disorders, neurodegenerative disease and diabetes (97,98). CircRNAs essentially originate from pre-miRNAs, are characterized by a covalently closed loop structure and regulate miRNAs expression through acting as miRNA sponges (99). These molecules carry some notable advantages compared to linear RNAs, being essentially more stable and expressed at high levels in paternal tissues (100).
Yan and colleagues (101) carried out next-generation sequencing (NGS) in placental villi of women with GDM and normal controls, and were capable of identifying as many as 48,270 circRNAs, 227 of which were found to be significantly up-regulated and 255 circRNAs significantly down-regulated in the GDM cohort. They could hence hypothesize that these circRNAs may play some important roles in the development of GDM (101). Wu et al. carried out an interesting study measuring six circRNAs (hsa_circRNA_0054633, hsa_circRNA_103410, hsa_circRNA_063981, hsa_circRNA_102682, hsa_circRNA_0018508, and hsa_circRNA_406918) in serum samples of 40 healthy pregnant women, 40 women with GDM during the second trimester of pregnancy, 65 controls and 65 GDM cases during the third trimester of pregnancy, as well as in placental tissues and cord blood of 20 GDM cases and 20 controls (102). Notably, circRNA_0054633 was found to be highly expressed in blood during the second and third trimesters. The expression was also high in the placenta, low in the cord blood (P<0.05) and was highly correlated with glycosylated hemoglobin (HbA1c) values levels in maternal blood samples. The assessment of this circRNA displayed a notable diagnostic performance in the second and third trimesters of pregnancy, placenta, and cord blood [area under the curve (AUC) of 0.79, 0.66, 0.75, and 0.78, respectively; all P<0.001] (102). Since circRNA_0054633 is involved in cell cycle progression and molecular catabolism (103), it was finally hypothesized that it may be also involved in regulating the proliferation of pancreatic β cells (104).
Very recently, Wang et al. explored the differential expression of circRNAs in the placentas of 30 GDM and 15 normal pregnant women (105). Among the 8,321 circRNAs identified in human placenta, three were found to be over-expressed and 43 down-regulated in GDM patients. By performing functional analysis of differentially expressed circRNAs, the authors concluded that these circRNAs may be active players in the pathogenesis of GDM since they are involved in advanced glycation end products-receptor for advanced glycation end products (AGE-RAGE) signaling pathway (106).
Conclusions
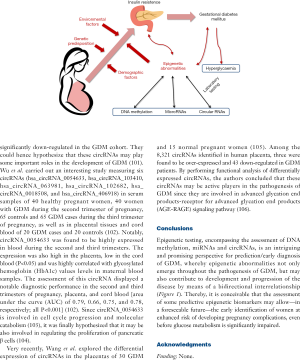
Epigenetic testing, encompassing the assessment of DNA methylation, miRNAs and circRNAs, is an intriguing and promising perspective for prediction/early diagnosis of GDM, whereby epigenetic abnormalities not only emerge throughout the pathogenesis of GDM, but may also contribute to development and progression of the disease by means of a bidirectional interrelationship (Figure 1). Thereby, it is conceivable that the assessment of some predictive epigenetic biomarkers may allow—in a foreseeable future—the early identification of women at enhanced risk of developing pregnancy complications, even before glucose metabolism is significantly impaired.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine for the series “Laboratory Medicine in Pregnancy”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm.2019.11.01). The series “Laboratory Medicine in Pregnancy” was commissioned by the editorial office without any funding or sponsorship. Giuseppe Lippi served as an unpaid Guest Editor of the series and serves as the unpaid Editor-in-Chief of Journal of Laboratory and Precision Medicine from November 2016 to October 2021. Martina Montagnana served as an unpaid Guest Editor of the series and serves as the unpaid Associate Editor-in-Chief of Journal of Laboratory and Precision Medicine from November 2016 to October 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- American Diabetes Association. 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care 2019;42:S13-28. [Crossref] [PubMed]
- Behboudi-Gandevani S, Amiri M, Bidhendi Yarandi R, et al. The impact of diagnostic criteria for gestational diabetes on its prevalence: a systematic review and meta-analysis. Diabetol Metab Syndr 2019;11:11. [Crossref] [PubMed]
- Ferrara A. Increasing prevalence of gestational diabetes mellitus: a public health perspective. Diabetes Care 2007;30:S141-6. [Crossref] [PubMed]
- Magee MS, Walden CE, Benedetti TJ, et al. Influence of diagnostic criteria on the incidence of gestational diabetes and perinatal morbidity. JAMA 1993;269:609-15. [Crossref] [PubMed]
- Yuen L, Wong VW. Gestational diabetes mellitus: challenges for different ethnic groups. World J Diabetes 2015;6:1024-32. [Crossref] [PubMed]
- Egan AM, Vellinga A, Harreiter J, et al. Epidemiology of gestational diabetes mellitus according to IADPSG/WHO 2013 criteria among obese pregnant women in Europe. Diabetologia 2017;60:1913-21. [Crossref] [PubMed]
- Plows JF, Stanley JL, Baker PN, et al. The pathophysiology of gestational diabetes mellitus. Int J Mol Sci 2018; [Crossref] [PubMed]
- Barbour LA, McCurdy CE, Hernandez TL, et al. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care 2007;30:S112-9. [Crossref] [PubMed]
- Wu Y, Sun G, Zhou X, et al. Pregnancy dietary cholesterol intake, major dietary cholesterol sources, and the risk of gestational diabetes mellitus: a prospective cohort study. Clin Nutr 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Murray-Davis B, Berger H, Melamed N, et al. Weight gain during pregnancy: does the antenatal care provider make a difference? A retrospective cohort study. CMAJ Open 2019;7:E283-93. [Crossref] [PubMed]
- Sapanont K, Sunsaneevithayakul P, Boriboonhirunsarn D. Relationship between ABO blood group and gestational diabetes mellitus. J Matern Fetal Neonatal Med 2019;1-5. [Crossref] [PubMed]
- Kleinberger JW, Maloney KA, Pollin TI. The genetic architecture of diabetes in pregnancy: implications for clinical practice. Am J Perinatol 2016;33:1319-26. [Crossref] [PubMed]
- Mirghani Dirar A, Doupis J. Gestational diabetes from A to Z. World J Diabetes 2017;8:489-511. [Crossref] [PubMed]
- Haller-Kikkatalo K, Uibo R. Clinical Recommendations for the use of islet cell autoantibodies to distinguish autoimmune and non-autoimmune gestational diabetes. Clin Rev Allergy Immunol 2016;50:23-33. [Crossref] [PubMed]
- Dennison RA, Fox RA, Ward RJ, et al. Women’s views on screening for type 2 diabetes after gestational diabetes: a systematic review, qualitative synthesis and recommendations for increasing uptake. Diabet Med 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Donnelly SR, Hinkle SN, Rawal S, et al. Prospective study of gestational diabetes and fatty liver scores 9 to 16 years after pregnancy. J Diabetes 2019;11:895-905. [Crossref] [PubMed]
- Lauenborg J, Mathiesen E, Hansen T, et al. The prevalence of the metabolic syndrome in a danish population of women with previous gestational diabetes mellitus is three-fold higher than in the general population. J Clin Endocrinol Metab 2005;90:4004-10. [Crossref] [PubMed]
- Kramer CK, Campbell S, Retnakaran R. Gestational diabetes and the risk of cardiovascular disease in women: a systematic review and meta-analysis. Diabetologia 2019;62:905-14. [Crossref] [PubMed]
- Mitanchez D, Yzydorczyk C, Siddeek B, et al. The offspring of the diabetic mother--short- and long-term implications. Best Pract Res Clin Obstet Gynaecol 2015;29:256-69. [Crossref] [PubMed]
- Li Y, Wang W, Zhang D. Maternal diabetes mellitus and risk of neonatal respiratory distress syndrome: a meta-analysis. Acta Diabetol 2019;56:729-40. [Crossref] [PubMed]
- Lowe WL Jr, Scholtens DM, Kuang A, et al. Hyperglycemia and Adverse Pregnancy Outcome Follow-up Study (HAPO FUS): maternal gestational diabetes mellitus and childhood glucose metabolism. Diabetes Care 2019;42:372-80. [Crossref] [PubMed]
- Li-Zhen L, Yun X, Xiao-Dong Z, et al. Evaluation of guidelines on the screening and diagnosis of gestational diabetes mellitus: systematic review. BMJ Open 2019;9:e023014 [Crossref] [PubMed]
- O’Sullivan JB, Mahan CM. Criteria for the oral glucose tolerance test in pregnancy. Diabetes 1964;13:278-85. [PubMed]
- O’Sullivan JB, Mahan CM, Charles D, et al. Screening criteria for high-risk gestational diabetic patients. Am J Obstet Gynecol 1973;116:895-900. [Crossref] [PubMed]
- National Diabetes Data Group. Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes 1979;28:1039-57. [Crossref] [PubMed]
- Carpenter MW, Coustan DR. Criteria for screening tests for gestational diabetes. Am J Obstet Gynecol 1982;144:768-73. [Crossref] [PubMed]
- Brown CJ, Dawson A, Dodds R, et al. Report of the pregnancy and neonatal care group. Diabet Med 1996;13:S43-53. [PubMed]
- World Health Organization. Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO consultation. Part 1, diagnosis and classification of diabetes mellitus. Geneva: World Health Organization, 1999.
- Metzger BE, Buchanan TA, Coustan DR, et al. Summary and recommendations of the fifth international workshop-conference on gestational diabetes mellitus. Diabetes Care 2007;30:S251-60. [Crossref] [PubMed]
- American Diabetes Association. Gestational diabetes mellitus. Diabetes Care 2000;23:S77-9. [PubMed]
- Metzger BE, Gabbe SG, Persson B, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010;33:676-82. [Crossref] [PubMed]
- World Health Organization. Diagnostic criteria and classification of hyperglycaemia first detected in pregnancy. 2013. Available online: http://apps.who.int/iris/bitstream/10665/85975/1/WHO_NMH_MND_13.2_eng.pdf
- National Institute for Health and Care Excellence. Diabetes in pregnancy: management from preconception to the postnatal period. 2015. Available online: https://www.nice.org.uk/guidance/ng3
- Hod M, Kapur A, Sacks DA, et al. The International Federation of Gynecology and Obstetrics (FIGO) Initiative on gestational diabetes mellitus: a pragmatic guide for diagnosis, management, and care. Int J Gynaecol Obstet 2015;131:S173-211. [Crossref]
- Committee on Practice Bulletins—Obstetrics. ACOG practice bulletin no. 190: gestational diabetes mellitus. Obstet Gynecol 2018;131:e49-64. [Crossref] [PubMed]
- Hod M, Kapur A, McIntyre HD, et al. Evidence in support of the International Association of Diabetes in Pregnancy study groups’ criteria for diagnosing gestational diabetes mellitus worldwide in 2019. Am J Obstet Gynecol 2019;221:109-16. [Crossref] [PubMed]
- HAPO Study Cooperative Research Group. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 2008;358:1991-2002. [Crossref] [PubMed]
- Agarwal MM, Dhatt GS, Shah SM. Gestational diabetes mellitus: simplifying the international association of diabetes and pregnancy diagnostic algorithm using fasting plasma glucose. Diabetes Care 2010;33:2018-20. [Crossref] [PubMed]
- Ogonowski J, Miazgowski T, Homa K, et al. Low predictive value of traditional risk factors in identifying women at risk for gestational diabetes. Acta Obstet Gynecol Scand 2007;86:1165-70. [Crossref] [PubMed]
- Diagnosi del diabete gestazionale. Linea-guida gravidanza fisiologica 2011;13:169-73. Available online: www.salute.gov.it/imgs/C_17_pubblicazioni_1436_allegato.pdf
- Bianchi C, de Gennaro G, Romano M, et al. Italian national guidelines for the screening of gestational diabetes: time for a critical appraisal? Nutr Metab Cardiovasc Dis 2017;27:717-22. [Crossref] [PubMed]
- Vitacolonna E, Succurro E, Lapolla A, et al. Guidelines for the screening and diagnosis of gestational diabetes in Italy from 2010 to 2019: critical issues and the potential for improvement. Acta Diabetol 2019;56:1159-67. [Crossref] [PubMed]
- Daly N, Flynn I, Carroll C, et al. Impact of implementing preanalytical laboratory standards on the diagnosis of gestational diabetes mellitus: a prospective observational study. Clin Chem 2016;62:387-91. [Crossref] [PubMed]
- Lippi G, Salvagno GL, Lampus S, et al. Impact of blood cell counts and volumes on glucose concentration in uncentrifuged serum and lithium-heparin blood tubes. Clin Chem Lab Med 2018;56:2125-31. [Crossref] [PubMed]
- Montagnana M, Lippi G. Overcoming preanalytical issues for diagnosing diabetes with fasting plasma glucose. Ann Transl Med 2017;5:257. [Crossref] [PubMed]
- Lippi G, Nybo M, Cadamuro J, et al. Blood glucose determination: effect of tube additives. Adv Clin Chem 2018;84:101-23. [Crossref] [PubMed]
- Daly N, Turner MJ. Laboratory diagnosis of gestational diabetes mellitus. BJOG 2016;123:1430-3. [Crossref] [PubMed]
- Jerram ST, Dang MN, Leslie RD. The role of epigenetics in type 1 diabetes. Curr Diab Rep 2017;17:89. [Crossref] [PubMed]
- van Dijk SJ, Tellam RL, Morrison JL, et al. Recent developments on the role of epigenetics in obesity and metabolic disease. Clin Epigenetics 2015;7:66. [Crossref] [PubMed]
- Dobosz AM, Dziewulska A, Dobrzyń A. Spotlight on epigenetics as a missing link between obesity and type 2 diabetes. Postepy Biochem 2018;64:157-65. [Crossref] [PubMed]
- Dhawan S, Natarajan R. Epigenetics and type 2 diabetes risk. Curr Diab Rep 2019;19:47. [Crossref] [PubMed]
- Pinel C, Prainsack B, McKevitt C. Markers as mediators: a review and synthesis of epigenetics literature. Biosocieties 2019;13:276-303. [Crossref] [PubMed]
- Lim DHK, Maher ER. DNA methylation: a form of epigenetic control of gene expression. Obstet Gynaecol 2010;12:37-42. [Crossref]
- Bansal A, Pinney SE. DNA methylation and its role in the pathogenesis of diabetes. Pediatr Diabetes 2017;18:167-77. [Crossref] [PubMed]
- Moen GH, Sommer C, Prasad RB, et al. Mechanisms in endocrinology: epigenetic modifications and gestational diabetes: a systematic review of published literature. Eur J Endocrinol 2017;176:R247-67. [Crossref] [PubMed]
- Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One 2010;5:e14398 [Crossref] [PubMed]
- Aagaard-Tillery KM, Grove K, Bishop J, et al. Developmental origins of disease and determinants of chromatin structure: Maternal diet modifies the primate fetal epigenome. J Mol Endocrinol 2008;41:91-102. [Crossref] [PubMed]
- Suter MA, Chen A, Burdine MS, et al. A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB J 2012;26:5106-14. [Crossref] [PubMed]
- Zhu Z, Chen X, Xiao Y, et al. Gestational diabetes mellitus alters DNA methylation profiles in pancreas of the offspring mice. J Diabetes Complications 2019;33:15-22. [Crossref] [PubMed]
- Rong C, Cui X, Chen J, et al. DNA methylation profiles in placenta and its association with gestational diabetes mellitus. Exp Clin Endocrinol Diabetes 2015;123:282-8. [Crossref] [PubMed]
- Nomura Y, Lambertini L, Rialdi A, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci 2014;21:131-7. [Crossref] [PubMed]
- Reichetzeder C, Dwi Putra SE, Pfab T, et al. Increased global placental DNA methylation levels are associated with gestational diabetes. Clin Epigenetics 2016;8:82. [Crossref] [PubMed]
- Deng X, Yang Y, Sun H, et al. Analysis of whole genome-wide methylation and gene expression profiles in visceral omental adipose tissue of pregnancies with gestational diabetes mellitus. J Chin Med Assoc 2018;81:623-30. [Crossref] [PubMed]
- Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem 2008;54:482-90. [Crossref] [PubMed]
- Dias S, Adam S, Van Wyk N, et al. Global DNA methylation profiling in peripheral blood cells of South African women with gestational diabetes mellitus. Biomarkers 2019;24:225-31. [Crossref] [PubMed]
- Enquobahrie DA, Moore A, Muhie S, et al. Early pregnancy maternal blood DNA methylation in repeat pregnancies and change in gestational diabetes mellitus status—a pilot study. Reprod Sci 2015;22:904-10. [Crossref] [PubMed]
- Wu P, Farrell WE, Haworth KE, et al. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics 2018;13:122-8. [Crossref] [PubMed]
- Kang J, Lee CN, Li HY, et al. Genome-wide DNA methylation variation in maternal and cord blood of gestational diabetes population. Diabetes Res Clin Pract 2017;132:127-36. [Crossref] [PubMed]
- Kurdyukov S, Bullock M. DNA methylation analysis: choosing the right method. Biology (Basel) 2016; [Crossref] [PubMed]
- Wróblewski A, Strycharz J, Świderska E, et al. Molecular insight into the interaction between epigenetics and leptin in metabolic disorders. Nutrients 2019; [Crossref] [PubMed]
- Bouchard L, Thibault S, Guay SP, et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care 2010;33:2436-41. [Crossref] [PubMed]
- Bouchard L, Hivert MF, Guay SP, et al. Placental adiponectin gene DNA methylation levels are associated with mothers’ blood glucose concentration. Diabetes 2012;61:1272-80. [Crossref] [PubMed]
- Kang J, Lee C-N, Li H-Y, et al. Association of interleukin-10 methylation levels with gestational diabetes in a Taiwanese population. Front Genet 2018;9:222. [Crossref] [PubMed]
- Zhang Y, Chen Y, Qu H, et al. Methylation of HIF3A promoter CpG islands contributes to insulin resistance in gestational diabetes mellitus. Mol Genet Genomic Med 2019;7:e00583 [Crossref] [PubMed]
- Main AM, Gillberg L, Jacobsen AL, et al. DNA methylation and gene expression of HIF3A: cross-tissue validation and associations with BMI and insulin resistance. Clin Epigenetics 2016;8:89. [Crossref] [PubMed]
- Morales-Prieto DM, Ospina-Prieto S, Schmidt A, et al. Elsevier Trophoblast Research Award Lecture: origin, evolution and future of placenta miRNAs. Placenta 2014;35:S39-45. [Crossref] [PubMed]
- Fu G, Brkić J, Hayder H, et al. MicroRNAs in human placental development and pregnancy complications. Int J Mol Sci 2013;14:5519-44. [Crossref] [PubMed]
- Gu Y, Sun J, Groome LJ, et al. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol Endocrinol Metab 2013;304:E836-43. [Crossref] [PubMed]
- Muralimanoharan S, Maloyan A, Myatt L. Mitochondrial function and glucose metabolism in the placenta with gestational diabetes mellitus: role of miR-143. Clin Sci (Lond) 2016;130:931-41. [Crossref] [PubMed]
- Li J, Song L, Zhou L, et al. A microRNA signature in gestational diabetes mellitus associated with risk of macrosomia. Cell Physiol Biochem 2015;37:243-52. [Crossref] [PubMed]
- Di Francesco L, Dovizio M, Trenti A, et al. Dysregulated post-transcriptional control of COX-2 gene expression in gestational diabetic endothelial cells. Br J Pharmacol 2015;172:4575-87. [Crossref] [PubMed]
- Floris I, Descamps B, Vardeu A, et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler Thromb Vasc Biol 2015;35:664-74. [Crossref] [PubMed]
- Zhao C, Zhang T, Shi Z, et al. MicroRNA-518d regulates PPARalpha protein expression in the placentas of females with gestational diabetes mellitus. Mol Med Rep 2014;9:2085-90. [Crossref] [PubMed]
- Shi Z, Zhao C, Guo X, et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology 2014;155:1982-90. [Crossref] [PubMed]
- Zhao C, Dong J, Jiang T, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One 2011;6:e23925 [Crossref] [PubMed]
- Zhu Y, Tian F, Li H, et al. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet 2015;130:49-53. [Crossref] [PubMed]
- Cao JL, Zhang L, Li J, et al. Up-regulation of miR-98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci Rep 2016;6:32268. [Crossref] [PubMed]
- Wander PL, Boyko EJ, Hevner K, et al. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res Clin Pract 2017;132:1-9. [Crossref] [PubMed]
- Pheiffer C, Dias S, Rheeder P, et al. Decreased expression of circulating mir-20a-5p in south african women with gestational diabetes mellitus. Mol Diagn Ther 2018;22:345-52. [Crossref] [PubMed]
- Tagoma A, Alnek K, Kirss A, et al. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018;672:137-42. [Crossref] [PubMed]
- He Y, Bai J, Liu P, et al. miR-494 protects pancreatic β-cell function by targeting PTEN in gestational diabetes mellitus. EXCLI J 2017;16:1297-307. [PubMed]
- Sebastiani G, Guarino E, Grieco GE, et al. Circulating microRNA (miRNA) expression profiling in plasma of patients with gestational diabetes mellitus reveals upregulation of miRNA miR-330-3p. Front Endocrinol (Lausanne) 2017;8:345. [Crossref] [PubMed]
- Stirm L, Huypens P, Sass S, et al. Maternal whole blood cell miRNA-340 is elevated in gestational diabetes and inversely regulated by glucose and insulin. Sci Rep 2018;8:1366. [Crossref] [PubMed]
- Lamadrid-Romero M, Solís KH, Cruz-Reséndiz MS, et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci Res 2018;130:8-22. [Crossref] [PubMed]
- Lippi G, Bassi A, Bovo C. The future of laboratory medicine in the era of precision medicine. J Lab Precis Med 2016;1:7. [Crossref]
- Lippi G, Simundic AM. The preanalytical phase in the era of high-throughput genetic testing. What the future holds. Diagnosis (Berl) 2019;6:73-4. [PubMed]
- Yang D, Yang K, Yang M. Circular RNA in aging and age-related diseases. Adv Exp Med Biol 2018;1086:17-35. [Crossref] [PubMed]
- Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Yan L, Feng J, Cheng F, et al. Circular RNA expression profiles in placental villi from women with gestational diabetes mellitus. Biochem Biophys Res Commun 2018;498:743-50. [Crossref] [PubMed]
- Wu H, Wu S, Zhu Y, et al. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin Epigenetics 2019;11:22. [Crossref] [PubMed]
- Zhao Z, Li X, Jian D, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol 2017;54:237-45. [Crossref] [PubMed]
- Rane SG, Reddy EP. Cell cycle control of pancreatic beta cell proliferation. Front Biosci 2000;5:D1-19. [Crossref] [PubMed]
- Wang H, She G, Zhou W, et al. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr J 2019;66:431-41. [Crossref] [PubMed]
- Goh SY, Cooper ME. Clinical review: the role of advanced glycation end products in progression and complications of diabetes. J Clin Endocrinol Metab 2008;93:1143-52. [Crossref] [PubMed]
Cite this article as: Montagnana M, Danese E, Lippi G. The current and future role of laboratory medicine for diagnosing gestational diabetes mellitus. J Lab Precis Med 2020;5:2.