Diagnostic accuracy of pleural effusion biomarkers for malignant pleural mesothelioma: a machine learning analysis
Introduction
Malignant pleural mesothelioma (MPM) is a lethal form of cancer with an inferior prognosis (1). The outcomes of MPM patients can be improved by accurate and timely diagnosis (2). More than half of MPM patients visit the hospital with complaints of chest pain and dyspnea (3). However, these symptoms are not specific to MPM, and MPM’s diagnosis is challenging for clinicians. According to the recent guideline released by the British Thoracic Society (BTS), diagnostic pleural aspiration, image-guided cutting needle biopsy, and thoracoscopy are recommended for diagnosing MPM (3). However, these diagnostic tools are invasive and are not available in all centers. Therefore, it is of great value to develop diagnostic tools with reduced invasiveness. Because pleural effusion (PE) is the most common sign for MPM, soluble biomarkers in PE have been proposed as a tool for diagnostic purposes. During the past decades, several PE biomarkers (4) [e.g., soluble mesothelin-related peptide (SMRP) (5), fibulin-3 (6,7), osteopontin (8,9) and cytokeratin 19 fragment (CYFRA 21-1) (4,10)] have been verified, and their diagnostic accuracy has been evaluated in various studies. However, no biomarker has sufficient diagnostic accuracy for MPM when used alone, according to the guidelines (3,11,12). Therefore, a multiple biomarker approach may be a promising strategy to improve the diagnostic accuracy for MPM.
Machine learning is a type of artificial intelligence. It allows computers to learn with data and build a data model to support a given task with various mathematical and algorithmic approaches (13,14). Machine learning has been used for diagnostic aims in various settings, especially in cancer diagnosis (15). However, the study investigating PE markers’ diagnostic accuracy for MPM with machine learning approaches is rare. In this study, we hypothesized that machine learning could improve PE biomarkers’ diagnostic accuracy for MPM. We present the following article in accordance with the Standards for Reporting of Diagnostic Accuracy Studies (STARD) reporting checklist (Tables S1,S2) (available at http://dx.doi.org/10.21037/jlpm-20-90).
Methods
Subjects
This is a post hoc analysis of a previous study (16). We obtained the data of this study at the Dryad online repository (http://datadryad.org/review?doi=doi:10.5061/dryad.fg0ft) (17). Briefly, the study mentioned above is a retrospective study performed in a hospital in Japan between September 2014 and August 2016. A total of 240 consecutive patients with undiagnosed PE were enrolled. The diagnostic accuracy of PE SMRP, carcinoembryonic antigen (CEA), and CYRFA 21-1 was assessed using receiver operating characteristic (ROC) curve analysis. Our study excluded the patients with missing values for SMRP, CEA, and CYRFA 21-1. This study was performed with shared data, and we conducted this study following the Declaration of Helsinki (as revised in 2013). Informed consent from the subjects and ethical approval from the authors’ institution were waived because the data used in this work are from the internet.
Statistical analysis
In this study, we used machine learning algorithms to evaluate the diagnostic accuracy of PE biomarkers. Briefly, the study cohort was randomly categorized into training and test cohorts. The training cohort was used for model building, and the test cohort was used for validation. The machine learning algorithms used in this study were: logistic regression model (18), linear discriminant analysis (LDA) (19), multivariate adaptive regression splines (MARS) (20), k-nearest neighbor (KNN) (21), support vector machine (SVM) (22), gradient boosting machine (GBM), and random forest. We used the ROC curve to evaluate the diagnostic accuracy of a single biomarker and the machine learning model (23). All analyses were performed with the caret package of R (version 4.0.1), and statistical significance was set at P<0.05.
Results
Characteristics of the subjects
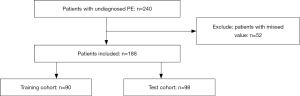
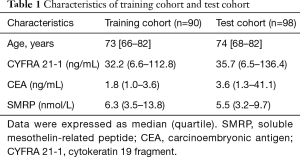
Figure 1 is a flowchart of the patient selection process. A total of 188 subjects, with 27 being diagnosed with MPM, were included in the present study. They were randomly categorized into training cohort (n=90) and test cohort (n=98). The characteristics of these two cohorts were listed in Table 1.
Full table
Evaluating the diagnostic accuracy of CEA^ SMRP^ and CYFRA 21-1 with machine learning algorithms
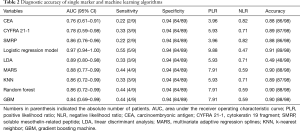
The diagnostic accuracy of single biomarker and machine learning algorithms was summarized in Table 2. When specificity was fixed at 0.94, the sensitivities of CEA, CYFRA 21-1, and SMRP were 0.22, 0.33, and 0.22, respectively. The logistic regression model increased the sensitivity (0.55) without decreasing specificity. Notably, the area under the ROC curve (AUC) of the logistic regression model was higher than that of a single marker and the other machine learning approaches.
Full table
Discussion
This study used machine learning approaches to evaluate the diagnostic accuracy of three conventional tumor markers, CEA, SMRP, and CYFRA 21-1, for MPM. The major finding of the present study is that combinational use of these three biomarkers with a logistic regression model can greatly improve the diagnostic accuracy, while other machine learning approaches had limited ability to improve the diagnostic accuracy. Therefore, the logistic regression model represents a potential machine learning algorithm to improve PE tumor markers’ diagnostic accuracy for MPM.
This is the second study investigating the diagnostic accuracy of PE tumor markers for MPM, to the best of our knowledge. In the previous study (24), the authors used a logic learning machine (LLM), KNN, artificial neural network (ANN), and decision tree (DT) to evaluate the diagnostic accuracy of PE CEA, SMRP, and CYFRA 21-1 for MPM. They found that the LLM, KNN, ANN, and DT sensitivities were 0.77, 0.56, 0.60, and 0.75, respectively, and the specificities were 0.91, 0.81, 0.86, and 0.92, respectively. Some new machine learning algorithms were used in our study, such as the logistic regression model and random forest. The specificities concluded in our study are generally higher than those of the previous study; however, sensitivities are relatively low. The inconsistency between the present and previous study may be due to the clinical characteristics of MPM and disease profiles of the study cohorts.
Sensitivity and specificity are two primary diagnostic test measures, but their clinical interpretation is not straightforward. The same degree of decrease in sensitivity and specificity can lead to a different number of missed diagnoses and misdiagnoses, which depends on the prevalence of the target disease in the study cohort. By contrast, the positive likelihood ratio (PLR) and negative likelihood ratio (NLR) represent two statistics that are not affected by the target disease’s prevalence (25). It is generally accepted that NLR <0.1 or PLR >10 provides strong evidence to rule out or rule in target disease (25). In our study, a PLR of 9.88 was observed in the logistic regression model, suggesting that the positive result of logistic regression is an evidence of MPM. Therefore, the logistic regression model represents a practical algorithm to rule in or out of MPM. AUC is a threshold independent indicator that reflects the overall diagnostic accuracy of an index test. The AUC of the logistic regression model is 0.97, indicating that the logistic regression model is a promising strategy for MPM diagnosis.
This study has two limitations. One limitation is the small sample size in the training and test cohort, and only three markers were considered. The other limitation is the retrospective design, which may introduce patients selection bias. In addition, the results of study have not been validated by other centers. Therefore, further prospective studies with large sample sizes are needed to validate the findings of this study.
Taken together, with three tumor markers in PE, we evaluated the diagnostic accuracy of some machine learning algorithms for MPM. Our results indicate that some machine learning algorithms, such as the logistic regression model, can improve PE tumor markers’ diagnostic accuracy. Give the small sample size and retrospective study design, and future studies are needed to validate our study’s findings.
Acknowledgments
Funding: This work was supported by the Natural and Science Foundation of Inner Mongolia Autonomous Region for Distinguished Young Scholars (2020JQ07).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Laboratory and Precision Medicine, for the series “Pleural Effusion Analysis”. This article has undergone external peer review.
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/jlpm-20-90
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jlpm-20-90
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jlpm-20-90). The series “Pleural Effusion Analysis” was commissioned by the editorial office without any funding or sponsorship. ZDH served as the unpaid Guest Editor of the series and serves as an unpaid executive editor of the Journal of Laboratory and Precision Medicine from Nov 2016 to Oct 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed with shared data, and the authors conducted this study following the Declaration of Helsinki (as revised in 2013). Informed consent from the subjects and ethical approval from the authors’ institution were waived because the data used in this work are from the internet.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tsao AS, Wistuba I, Roth JA, et al. Malignant pleural mesothelioma. J Clin Oncol 2009;27:2081-90. [Crossref] [PubMed]
- Ismail-Khan R, Robinson LA, Williams CC Jr, et al. Malignant pleural mesothelioma: a comprehensive review. Cancer Control 2006;13:255-63. [Crossref] [PubMed]
- Woolhouse I, Bishop L, Darlison L, et al. British Thoracic Society Guideline for the investigation and management of malignant pleural mesothelioma. Thorax 2018;73:i1-30. [Crossref] [PubMed]
- Gillezeau CN, van Gerwen M, Ramos J, et al. Biomarkers for malignant pleural mesothelioma: a meta-analysis. Carcinogenesis 2019;40:1320-31. [Crossref] [PubMed]
- Cui A, Jin XG, Zhai K, et al. Diagnostic values of soluble mesothelin-related peptides for malignant pleural mesothelioma: updated meta-analysis. BMJ Open 2014;4:e004145. [Crossref] [PubMed]
- Ren R, Yin P, Zhang Y, et al. Diagnostic value of fibulin-3 for malignant pleural mesothelioma: a systematic review and meta-analysis. Oncotarget 2016;7:84851-9. [Crossref] [PubMed]
- Ledda C, Caltabiano R, Vella F, et al. Fibulin-3 as biomarker of malignant mesothelioma. Biomark Med 2019;13:875-86. [Crossref] [PubMed]
- Hu ZD, Liu XF, Liu XC, et al. Diagnostic accuracy of osteopontin for malignant pleural mesothelioma: a systematic review and meta-analysis. Clin Chim Acta 2014;433:44-8. [Crossref] [PubMed]
- Lin H, Shen YC, Long HY, et al. Performance of osteopontin in the diagnosis of malignant pleural mesothelioma: a meta-analysis. Int J Clin Exp Med 2014;7:1289-96. [PubMed]
- Suzuki H, Hirashima T, Kobayashi M, et al. Cytokeratin 19 fragment/carcinoembryonic antigen ratio in pleural effusion is a useful marker for detecting malignant pleural mesothelioma. Anticancer Res 2010;30:4343-6. [PubMed]
- Kindler HL, Ismaila N, Armato SG 3rd, et al. Treatment of malignant pleural mesothelioma: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2018;36:1343-73. [Crossref] [PubMed]
- Scherpereel A, Opitz I, Berghmans T, et al. ERS/ESTS/EACTS/ESTRO guidelines for the management of malignant pleural mesothelioma. Eur Respir J 2020;55:1900953. [Crossref] [PubMed]
- Cabitza F, Banfi G. Machine learning in laboratory medicine: waiting for the flood? Clin Chem Lab Med 2018;56:516-24. [Crossref] [PubMed]
- Gruson D, Helleputte T, Rousseau P, et al. Data science, artificial intelligence, and machine learning: opportunities for laboratory medicine and the value of positive regulation. Clin Biochem 2019;69:1-7. [Crossref] [PubMed]
- Akazawa M, Hashimoto K. Artificial intelligence in ovarian cancer diagnosis. Anticancer Res 2020;40:4795-800. [Crossref] [PubMed]
- Otoshi T, Kataoka Y, Ikegaki S, et al. Pleural effusion biomarkers and computed tomography findings in diagnosing malignant pleural mesothelioma: a retrospective study in a single center. PLoS One 2017;12:e0185850. [Crossref] [PubMed]
- Miller GW. Making data accessible: the dryad experience. Toxicol Sci 2016;149:2-3. [Crossref] [PubMed]
- Zhang Z. Model building strategy for logistic regression: purposeful selection. Ann Transl Med 2016;4:111. [Crossref] [PubMed]
- Chan YH. Biostatistics 303. Discriminant analysis. Singapore Med J 2005;46:54-61. [PubMed]
- Friedman JH, Roosen CB. An introduction to multivariate adaptive regression splines. Stat Methods Med Res 1995;4:197-217. [Crossref] [PubMed]
- Zhang Z. Introduction to machine learning: k-nearest neighbors. Ann Transl Med 2016;4:218. [Crossref] [PubMed]
- Noble WS. What is a support vector machine? Nat Biotechnol 2006;24:1565-7. [Crossref] [PubMed]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Parodi S, Filiberti R, Marroni P, et al. Differential diagnosis of pleural mesothelioma using Logic Learning Machine. BMC Bioinformatics 2015;16 Suppl 9:S3. [Crossref] [PubMed]
- Deeks JJ, Altman DG. Diagnostic tests 4: likelihood ratios. BMJ 2004;329:168-9. [Crossref] [PubMed]
Cite this article as: Niu Y, Hu ZD. Diagnostic accuracy of pleural effusion biomarkers for malignant pleural mesothelioma: a machine learning analysis. J Lab Precis Med 2021;6:4.