Leukocyte differential and cell population data in the evaluation of SARS-CoV-2 and other infections
Highlight box
Key findings
• WBC differential and CPD reported by Mindray BC 6800 Plus analyzer seem reliable parameters for the initial evaluation of patients with fever of unknown origin.
What is known and what is new?
• WBC differential and CPD present typical values in bacterial, viral and SARS-CoV-2 infections.
• Neutrophilia and lymphopenia are common findings in COVID-19 patients.
• Using unsupervised K-means clustering method COVID-19 can be efficiently recognized: patients can be classified into distinct groups according to the etiology of the infection.
What is the implication, and what should change now?
• The use of advanced parameters, such as CPD, could be a way to improve the algorithms combining standard and research parameters.
Introduction
Coronavirus disease 2019 (COVID-19) can begin with flu-like symptoms. The main clinical signs are fever, coughing, myalgia, and headache. These might be followed by respiratory distress, chest pain and fatigue, suggestive of the massive involvement of the lungs as the principal organ, leading to severe pneumonia (1). The disease progression and severity can be classified into stages I–III. The main analytical manifestation of stage III is the increase in the concentration of systemic inflammation markers (2).
The current gold standard for etiological assessment is real-time reverse transcription-polymerase chain reaction (rRT-PCR) using oral or nasopharyngeal swab specimens (3). This technique is time-consuming and overwhelmed by large numbers of patients, clinical laboratories need diagnostic approaches that can rapidly and accurately aid to identify SARS-Cov-2 infected individuals.
Abnormalities in leukocyte and coagulation can be detected even in the early phase of the infection (4-8).
Complete blood count (CBC) and leukocyte differential present specific features in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected patients, which could be useful for screening and prognosis of the outcome of patients. Neutrophilia, lymphopenia, thrombocytopenia, and systemic inflammation markers, such as neutrophil-to-lymphocyte ratio (NLR), show the most significant quantitative alterations, and are recognized as predictors of disease severity (9-13).
Regarding infections with other etiologies, leukocyte and absolute neutrophil count, percentage of neutrophils, and especially, increases in immature neutrophils have been used as a laboratory test representing acute bacterial infections, while lymphocytosis and atypical lymphocyte can be found in increased numbers in viral illnesses.
The modern generation of Hematology counters applies innovative analytical principles. New morphometric parameters called Cell Population Data (CPD) are research parameters reported along with the CBC. The first company to introduce these parameters was Beckman Coulter Inc. (Brea, California, USA) and later Sysmex Corporation (Kobe, Japan). Recently, Mindray Medical International Co. (Shenzhen, China) has launched the BC-6800 Plus analyser providing those CPD parameters.
Diverse studies on CPD values have reported changes in morphology of leukocytes (volume, cytoplasmic characteristics, content in nucleic acids) in response to acute bacterial or viral infections and sepsis (14).
Alterations in neutrophil morphology (size, shape and composition), mechanics (deformability) and motility (chemotaxis and migration) have been observed during infection. The structure of the cytoplasm in neutrophils exhibits granulation with the nucleic acid content increasing as a result of cytokine synthesis. Atypical lymphocytes can be detected in COVID-19 patients. Usually, these cells present with heterogeneous morphological features including a larger size, a round nucleus often with a large nucleolus, and abundant, deeply basophilic cytoplasm. The volume of monocytes is also altered in activated cells.
These morphological changes can be measured quantitatively using the CPD parameters (14-26).
Diverse articles have been published since the pandemic outbreak. CPD has been evaluated for the early detection of the disease, monitoring and prognosis of its potential severity.
Here, we explore the leukocyte differential and CPD parameters reported by the Mindray BC-6800 Plus analyser to classify patients into distinct groups according to the etiology of the infection at admission to the Emergency Department. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-47/rc).
Methods
Patients
This prospective observational study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), after receiving approval from the Comité de Ética de la Investigación de Euskadi CEIE (Regional Ethics Committee, No. PI2019090, 17th July 2019). Written informed consent was obtained from the patients when recruited for the study.
The recruitment period for the study group was from 1st December 2020 to 30th January 2021, included consecutive patients with suspected infection admitted to the Emergency Department.
The criterion for inclusion in the validation group was the same as for the study group: The recruitment period was 1st February to 30th November 2021.
Age, gender, clinical symptoms, underlying diseases and laboratory results at admission were recorded and retrieved from the Hospital and Laboratory Information Systems.
The CBC and CPD parameters were analysed in a Mindray BC-6800 Plus analyser (Mindray Diagnostics, Shenzhen, China). The manufacturer’s recommendations were used for the calibration and control procedures.
Patients with COVID-19 were diagnosed using the current standards and on the basis of positive results of RT-PCR for SARS-CoV-2 in throat swab specimens. Positive cultures proved bacterial infections, and molecular tests or positive serology confirmed other viral infections.
Instrument characteristics
The Mindray BC-6800 Plus system uses sheathed flow impedance combined with laser optics and fluorescence for the measurement of peripheral blood cells.
Leukocytes (WBC) are counted and classified in analytical channels with lysing and staining reagents. The optical signals are detected by channel-specific detectors. Three signals are reported for each leukocyte forward scatter, side scatter, and fluorescence related to cell size, cytoplasmic granules and nuclear characteristics, respectively (S-Cube technology) (27).
All measurements are combined in scattergrams, which provide a visual representation of the leukocyte differential showing clusters according to the morphology. The optical signals along X-axis (side scatter) are proportional to the internal complexity; fluorescence along Y-axis correlates with the nucleic acid content, while forward scatter (Z-axis) is related to cell size (Figure 1).
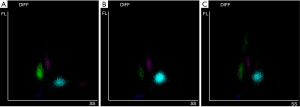
Each cell can be described by those 3 optical signals appearing in a scattergram (Figure 1). The coordinates to compose this scatterplot are the morphometric parameters, so called CPD (Table 1).
Table 1
| CPD | Description of CPD | Information of cell morphology |
|---|---|---|
| X axis | ||
| Neu X | Neutrophil complexity | Granules, vacuoles and other cytoplasmic inclusions |
| Lym X | Lymphocyte complexity | – |
| Mon X | Monocyte complexity | – |
| Y axis | – | Cellular DNA and RNA |
| Neu Y | Neutrophil fluorescence intensity | – |
| Lym Y | Lymphocyte fluorescence intensity | – |
| Mon Y | Monocyte fluorescence intensity | – |
| Z axis | – | Abnormal sized cells after staining |
| Neu Z | Neutrophil size | – |
| Lym Z | Lymphocyte size | – |
| Mon Z | Monocyte size | – |
CPD, cell population data.
CPD are research parameters allowing a detailed study of the cells and the detection of morphological variations in response to stimuli, such as infections of diverse etiologies (Table 1).
Activated leukocytes, in contrast with non-reactive cells, express signaling receptors which make their membranes more susceptible to perforation and the action of fluorescence dyes.
Therefore, CPD can offer valuable information regarding activation status of the leukocytes.
Statistical analysis
Kolmogorv-Smirnov test was applied to verify skewed or normal distributions. A preliminary exploratory data analysis was performed. For continuous variables, medians and interquartile range (IQR), frequencies and percentages for categorical data were calculated. Kruskal-Wallis test was applied in order to detect statistical differences among patients with infections of diverse etiologies.
No missing data imputation techniques were applied.
Next step was to test the hypothesis that infections of divers aetiologies could be distinguished into distinct groups based on their analytical status using the k-means unsupervised clustering method.
Patients were divided into two subsets, the study and the validation groups. The non-parametric Wilcoxon test (for continuous variables) and the χ2-test or Fisher’s exact test as appropriate (for categorical ones) were performed to assess the homogeneity of demographic data and analytical features.
K-means is a statistical technique which maximizes the similarity within each cluster and minimizes the similarities between clusters. Firstly, all the assessed CPD parameters were scaled and normalized by converting into z-scores. After that, the above mentioned statistical method was applied.
To obtain the optimal cluster number, a set of CBC and CPD were computed varying all the combinations of number of clusters, in order to decide the best clustering scheme. Additionally, the Principal component analysis (PCA) was used to validate to the choice of the optimal number of clusters, and to plot data points according to the obtained optimal principal components. This was performed in both the study and the validation groups. A confusion matrix in the validation set as computed to assess the performance of the model.
A P value <0.05 was deemed to be statistically significant. SAS 9.4 and R 3.4.1 were used to calculate the statistical procedures.
Results
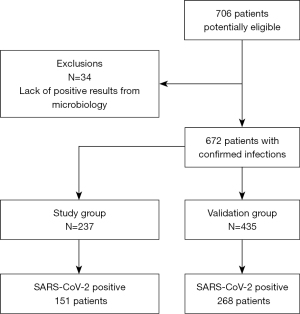
Seven hundred and six patients were potentially eligible. Thirty-four of them were excluded due to inconclusive microbiology results. Finally, 672 patients with suspected infection were recruited at admission to the hospital. Eighty-five percent of the patients were admitted to general awards and 15% to the Intensive Care Unit.
The study group included 237 patients; 151 of them (63.7%) suffered COVID-19. Fifty-three had bacterial infections, including respiratory infections and pneumonia, and 33 had other viral infections (respiratory infections, influenza, Epstein Barr).
The reliability of the model was evaluated in a validation group of 435 patients, 268 COVID-19 (61.6%) and 167 non-COVID-19, among which 51 had other virus and 116 suffered bacterial infections (Figure 2).
Table 2 displays the clinical features of the COVID-19 patients. Table 3 shows the analytical data in the study group, and Table 4 summarizes statistics of leukocytes (absolute counts and differential) and CPD, comparing both cohorts; mean ages were 63 years old [standard deviation (SD): 14.3] for study group and 64 years old (SD: 16.6) for validation group, P=0.077.
Table 2
| Patient characteristics | Data |
|---|---|
| Age, years, mean [25th–75th quartiles] | 64 [37–85] |
| Gender, male | 63.4% |
| Comorbidities | |
| Hypertension | 44.7% |
| Hypercholesterol | 38.47% |
| Diabetes mellitus | 17.2% |
| Cardiovascular disease | 14.63% |
| Chronic kidney disease | 7.95% |
| Pulmonary disease | 3.9% |
| Chronic Liver disease | 3.18% |
| Clinical characteristics | |
| Carlson index | |
| 0 | 52.78% |
| 1 | 24.01% |
| 2 | 10.33% |
| >2 | 12.88% |
| Intensive care unit | 17.38% |
| Mortality 30 days | 6.68% |
| Ward >12 days | 31.9% |
| Invasive respiratory support | 10.81% |
| 30-day readmission | 6.2% |
| Clinical signs at emergency department | |
| Fever | 100% |
| Dry cough | 78% |
| Fatigue | 43% |
| Dysnea | 33% |
| Laboratory data at emergency department, median [25th–75th quartiles] | |
| Creatinine, mg/dL | 1.42 [0.75–1.09] |
| Alanine aminotransferase, U/L | 40 [19–140] |
| Lactate dehydrogenase, U/L | 337 [206–564] |
| C reactive protein, mg/L | 97 [10.1–278] |
| Ferritin, μg/L | 1,153 [256–2,536] |
COVID-19, coronavirus disease 2019.
Table 3
| Parameters | Bacterial (n=53) | Other virus (n=33) | COVID-19 (n=151) | P value |
|---|---|---|---|---|
| WBC, 109/L | 12.55 (8.82, 16.8) | 8.87 (5.77, 12.3) | 7.19 (5.13, 9.6) | <0.001 |
| Neutrophils, 109/L | 9.92 (5.95, 14.0) | 3.68 (2.50, 4.6) | 5.69 (3.87, 7.8) | <0.001 |
| Lymphocytes, 109/L | 1.38 (0.95, 1.9) | 4.13 (2.78, 5.3) | 1.00 (0.68, 1.4) | <0.001 |
| Monocytes, 109/L | 0.69 (0.45, 1.0) | 0.64 (0.51, 0.8) | 0.26 (0.00, 0.5) | <0.001 |
| Platelets, 109/L | 194 (114, 359) | 251 (212, 370) | 222 (176, 302) | 0.0286 |
| NLR | 8.05 (4.13, 12.6) | 0.90 (0.47, 1.3) | 5.43 (3.79, 8.0) | <0.001 |
| Neu X | 376 (351, 411) | 334 (317, 358) | 346 (322, 372) | <0.001 |
| Neu Y | 451 (419, 469) | 409 (399, 442) | 428 (412, 448) | <0.001 |
| Neu Z | 1,821 (1,754, 1,866) | 1,811 (1,781, 1,859) | 1,808 (1,748, 1,876) | 0.898 |
| Lym X | 90 (86, 97) | 85 (80, 89) | 88 (85, 92) | 0.003 |
| Lym Y | 664 (630, 714) | 649 (624, 684) | 650 (627, 682) | 0.182 |
| Lym Z | 957 (936, 977) | 953 (932, 976) | 960 (945, 981) | 0.202 |
| Mon X | 203 (194, 218) | 196 (188, 200) | 200 (193, 209) | 0.001 |
| Mon Y | 917 (876, 976) | 960 (892, 994) | 933 (887, 990) | 0.443 |
| Mon Z | 1,315 (1,273, 1,341) | 1,290 (1,278, 1,310) | 1,343 (1,290, 1,352) | 0.01 |
| IG, 109/L | 0.07 (0.02, 0.3) | 0.01 (0.01, 0.0) | 0.01 (0.00, 0.0) | <0.001 |
The values are expressed as medians and quartiles (P25, P75) due to the skewed distribution of the data. Cell population data are reported in arbitrary optical units. COVID-19, coronavirus disease 2019; P, Kruskal-Wallis test; WBC, leukocyte count; NLR, neutrophil/lymphocyte ratio; Neu X, neutrophils complexity; Neu Y, neutrophils fluorescence; Neu Z, neutrophils size; Lym X, lymphocytes complexity; Lym Y, lymphocytes fluorescence; Lym Z, lymphocytes size; Mon X, monocytes complexity; Mon Y, monocytes fluorescence; Mon Z, monocytes size; IG, immature neutrophils count.
Table 4
| Parameters | Study group (n=237) | Validation group (n=435) | P value |
|---|---|---|---|
| WBC, 109/L | 8.11 (5.70, 11.92) | 8.17 (6.01, 11.70) | 0.05 |
| Lymphocytes, 109/L | 1.18 (0.70, 1.89) | 1.30 (0.72, 2.22) | 0.05 |
| Monocytes, 109/L | 0.40 (0.12, 0.66) | 0.45 (0.15, 0.68) | 0.06 |
| Neutrophils, 109/L | 5.86 (3.80, 9.01) | 5.87 (3.66, 9.03) | 0.44 |
| IG, 109/L | 0.01 (0.00, 0.04) | 0.00 (0.01, 0.05) | 0.04 |
| Neu X | 351 (323, 383) | 355 (328, 389) | 0.06 |
| Neu Y | 432 (409, 452) | 444 (430, 461) | 0.05 |
| Neu Z | 1,810 (1,754, 1,868) | 1,816 (1,756, 1,877) | 0.65 |
| Lym X | 88 (84, 93) | 92 (86, 96) | 0.21 |
| Lym Y | 651 (627, 688) | 659 (631, 701) | 0.07 |
| Lym Z | 957 (942, 979) | 964 (948, 984) | 0.11 |
| Mon X | 200(193, 209) | 204 (195, 214) | 0.33 |
| Mon Y | 933 (887, 988) | 952 (889, 1,006) | 0.38 |
| Mon Z | 1,301 (1,272, 1,331) | 1,299 (1,262, 1,328) | 0.91 |
| NLR | 4.93 (3.17, 8.6) | 4.96 (2.99, 8.53) | 0.05 |
The values are expressed as medians and quartiles (P25, P75). Cell population data are reported in arbitrary optical units. P, non-parametric Wilcoxon test; WBC, leukocyte count; Neu X, neutrophils complexity; Neu Y, neutrophils fluorescence; Neu Z, neutrophils size; Lym X, lymphocytes complexity; Lym Y, lymphocytes fluorescence; Lym Z, lymphocytes size; Mon X, monocytes complexity; Mon Y, monocytes fluorescence; Mon Z, monocytes size; IG, immature neutrophils count; NLR, neutrophil/lymphocyte ratio.
The results in the group of bacterial infections reflected the activation of the innate immunity system, showing the highest values in WBC, neutrophil counts, NLR and left shift. The neutrophil CPD parameters Neu X, Neu Y and Neu Z showed specific activation of this subset in terms of cytoplasmic granules, high RNA content and increased volume. Lymphocytosis and low NLR was observed in the viral group.
COVID-19 showed the characteristic neutrophilia, lymphopenia and high NLR while Neu X and Neu Y presented intermediate values and Mon Z is increased compared to infections of other etiologies.
The K-means analysis indicated an optimal clustering of two groups to classify the patients.
It was obtained from the combination of the CBC and CPD data within the K-means algorithm. PCA analysis enabled cluster validity to be assessed and PCA showed that almost 45.71% of the variability in the data was explained by the two components in the study group (Figure 3).
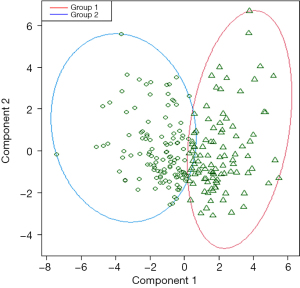
In this study group (237 patients), Cluster 1 was composed of 147 patients, 105 of them were COVID-19 patients and the other 90 patients were assigned to Cluster 2, which included 63.6% of other viral infections and 43.4% of bacterial infections.
The model was applied to the validation set; PCA analysis showed that almost 43.64% of the variability in the data was explained by the two components.
Table 5 displays the classification matrix of the observed infection groups against the predicted clusters. Cluster 1 included 91.4% of the COVID-19 patients and 60.5% of non-COVID-19 patients were assigned to Cluster 2, notably 100% of viral infections.
Table 5
| Infection group | Predicted cluster | Total (n=435) | |
|---|---|---|---|
| Cluster 1 | Cluster 2 | ||
| Bacteria, n (%) | 66 (56.90) | 50 (43.10) | 116 (26.67) |
| Virus, n (%) | 0 (0.00) | 51 (100.00) | 51 (11.72) |
| COVID-19, n (%) | 245 (91.42) | 23 (8.58) | 268 (61.60) |
Absolute number of patients (in brackets percentages) in each group and in the 2 clusters. COVID-19, coronavirus disease 2019.
Higher values in neutrophil counts, NLR, Neu X, Neu Y and Mon Z characterized Cluster 1 in contrast to the higher lymphocytes counts found in Cluster 2 (P<0.05).
Discussion
The pathology of COVID-19 results from an imbalance in immune response caused by dysregulation of the adaptive immune system (28). Since the pandemic outbreak, the hematological abnormalities have been reported in SARS-CoV-2 infected patients of 2020 (9,29).
The WBC differential and CPD results could help discriminate between the pathogens causing the infection (viral, bacterial) and the types of immune response (early innate, cellular or humoral). COVID-19 patients present a unique set of characteristics, as shown in the present study, such as neutrophilia, most commonly caused by an acute bacterial infection, and lymphopenia while lymphocytosis is typical for other viral infections (30,31). In the present study, the combination of CBC and CPD parameters allows to recognize COVID-19 patients in a distinct cluster in contrast to patients with fever or other etiologies.
Neutrophils and monocytes are among the first responses to an infection, and their biology appears considerably perturbed in patients with SARS-COV-2 infection (9,32).
At presentation, our COVID-19 patients showed alterations in both lineages, with increased internal complexity and nucleic acid content in neutrophils (Neu X, Neu Y) and a rise in monocyte volume (Mon Z). Our preliminary study using the Mindray BC-6800 Plus analyzer also showed that WBC, neutrophil counts and fluorescence (Neu Y) performed with high accuracy for predicting COVID-19 (33).
NLR is a valuable marker of inflammation; our results are in good agreement with previous reports of high NLR inCOVID-19 patients (23,33). Moreover, NLR is an independent indicator of the clinical outcomes (34,35).
A study has suggested that SARS-CoV-2 disrupts lymphocyte equilibrium and can promote the initial hyperactivation followed by the rapid exhaustion of cytotoxic CD8+ T-cells (36); this subset is hyper-reactive with increased cytotoxicity. Thus, an inflammation caused by SARS-CoV-2 increases the NLR and the reactive lymphocytes reflect this activation, allowing an insight into the overall inflammatory status (37).
COVID-19 and other infections can have a similar clinical presentation. Nevertheless, the diverse pathogens produce distinctive immune responses, reflected by leukocyte differentials; in addition, CPD add useful information on the activation of the leukocyte subsets. All those data represent the immune system status, so we hypothesized whether a leukocyte typical pattern of COVID-19, different form other infections, could be assessed.
For this purpose, we applied k means analysis which divides data into groups (clusters) in such a way that points in the same cluster are more similar to each other than those in other clusters where data are organized around the centroids.
In the study group, the classification matrix included two clusters: 89.2% of the COVID-19 patients were assigned to Cluster 1 and 53.5% of bacterial and other virus infections were included in Cluster 2.
In the validation group, 346 patients (79.5%) were correctly divided according to COVID-19 status. Cluster 1 included 91.4% of the COVID-19 patients, and 60.5% of non-COVID-19 patients were assigned to Cluster 2, notably 100% of viral infections.
Our study has some limitations. It is a single-centre study; the preliminary results must be validated in a multicentric study recruiting more patients. We aimed to improve the rapid evaluation of a SARS-CoV-2 infection at the Emergency Department; thus, the present study lacks data on asymptomatic patients and children.
The accurate evaluation of laboratory indicators at the onset of COVID-19 can help clinicians decide on the treatment. Abnormalities in routine tests, particularly in CBC, can warn of COVID-19 infection in a fast, practical and economical way (32).
Confirmatory diagnostic tests are time-consuming, so rapid indicators at triage in the emergency departments would be beneficial when SARS-CoV-2 infection is suspected for a better “watchful waiting” systems for such patients (38).
The CBC, the laboratory test most frequently ordered by emergency physicians, can be easily used for this purpose. The use of advanced parameters, such as CPD, could be a way to improve the algorithms combining standard and research parameters; Neutrophil counts, NLR, Neu X, Neu Y and Mon Z values could trigger an alarm or add a comment of “suspected COVID19” to the clinicians, which is clinically relevant information.
CPD from Beckman Coulter and Sysmex counters have been investigated, and the results obtained indicate that CPD largely depend on the analyser; this fact reveals a weakness of these “upgraded” WBC differentials: CPD from analyzers of different brands are not necessarily comparable.
The transferability and harmonisation must be ensured in order to introduce CPD in daily clinical practice.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-47/rc
Data Sharing Statement: Available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-47/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jlpm.amegroups.com/article/view/10.21037/jlpm-22-47/coif). EU serves as an unpaid editorial board member of Journal of Laboratory and Precision Medicine from October 2019 to November 2023. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This prospective observational study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), after receiving approval from the Comité de Ética de la Investigación de Euskadi CEIE (Regional Ethics Committee, No. PI2019090, 17 July 2019). Written informed consent was obtained from the patients when they were recruited for the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: A clinical-therapeutic staging proposal. J Heart Lung Transplant 2020;39:405-7. [Crossref] [PubMed]
- Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; [Crossref] [PubMed]
- Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;130:2620-9. [Crossref] [PubMed]
- Luan RS, Wang X, Sun X, et al. Epidemiology, Treatment, and Epidemic Prevention and Control of the Coronavirus Disease 2019: a Review. Sichuan Da Xue Xue Bao Yi Xue Ban 2020;51:131-8. [PubMed]
- Wu C, Chen X, Cai Y, et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med 2020;180:934-43. [Crossref] [PubMed]
- Kong J, Wang T, Di Z, et al. Analysis of hematological indexes of COVID-19 patients from fever clinics in Suzhou, China. Int J Lab Hematol 2020;42:e204-6. [Crossref] [PubMed]
- Singh K, Mittal S, Gollapudi S, et al. A meta-analysis of SARS-CoV-2 patients identifies the combinatorial significance of D-dimer, C-reactive protein, lymphocyte, and neutrophil values as a predictor of disease severity. Int J Lab Hematol 2021;43:324-8. [Crossref] [PubMed]
- Frater JL, Zini G, d'Onofrio G, et al. COVID-19 and the clinical hematology laboratory. Int J Lab Hematol 2020;42:11-8. [Crossref] [PubMed]
- Fan BE, Chong VCL, Chan SSW, et al. Hematologic parameters in patients with COVID-19 infection. Am J Hematol 2020;95:E131-4. [PubMed]
- Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta 2020;506:145-8. [Crossref] [PubMed]
- Rolla R, Vidali M, Puricelli C, et al. Reduced activity of B lymphocytes, recognised by Sysmex XN-2000™ haematology analyser, predicts mortality in patients with coronavirus disease 2019. Int J Lab Hematol 2021;43:e5-8. [Crossref] [PubMed]
- Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther 2020;5:33. [Crossref] [PubMed]
- Urrechaga E. Reviewing the value of leukocytes cell population data (CPD) in the management of sepsis. Ann Transl Med 2020;8:953. [Crossref] [PubMed]
- Zhu Y, Cao X, Lu Y, et al. Lymphocyte cell population as a potential hematological index for early diagnosis of COVID-19. Cell Mol Biol (Noisy-le-grand) 2020;66:202-6. [Crossref] [PubMed]
- Ognibene A, Lorubbio M, Magliocca P, et al. Elevated monocyte distribution width in COVID-19 patients: The contribution of the novel sepsis indicator. Clin Chim Acta 2020;509:22-4. [Crossref] [PubMed]
- Naoum FA, Ruiz ALZ, Martin FHO, et al. Diagnostic and prognostic utility of WBC counts and cell population data in patients with COVID-19. Int J Lab Hematol 2021;43:124-8. [Crossref] [PubMed]
- Zeng X, Xing H, Wei Y, et al. Monocyte volumetric parameters and lymph index are increased in SARS-CoV-2 infection. Int J Lab Hematol 2020;42:e266-9. [Crossref] [PubMed]
- Vasse M, Ballester MC, Ayaka D, et al. Interest of the cellular population data analysis as an aid in the early diagnosis of SARS-CoV-2 infection. Int J Lab Hematol 2021;43:116-22. [Crossref] [PubMed]
- Lippi G, Sanchis-Gomar F, Henry BM. Pooled analysis of monocyte distribution width in subjects with SARS-CoV-2 infection. Int J Lab Hematol 2021;43:O161-3. [Crossref] [PubMed]
- Linssen J, Ermens A, Berrevoets M, et al. A novel haemocytometric COVID-19 prognostic score developed and validated in an observational multicentre European hospital-based study. Elife 2020; [PubMed]
- Martens RJH, van Adrichem AJ, Mattheij NJA, et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the sysmex XN-10 hematology analyzer. Clin Chem Lab Med 2020;59:783-93. [Crossref] [PubMed]
- Urrechaga E, Aguirre U, España PP, et al. Complete blood counts and cell population data from Sysmex XN analyser in the detection of SARS-CoV-2 infection. Clin Chem Lab Med 2020;59:e57-60. [Crossref] [PubMed]
- Lapić I, Brenčić T, Rogić D, et al. Cell population data: Could a routine hematology analyzer aid in the differential diagnosis of COVID-19? Int J Lab Hematol 2021;43:e64-7. [Crossref] [PubMed]
- Harte JV, Mykytiv V. A panhaemocytometric approach to COVID-19: a retrospective study on the importance of monocyte and neutrophil population data on Sysmex XN-series analysers. Clin Chem Lab Med 2021;59:e169-72. [Crossref] [PubMed]
- Martens RJH, Leers MPG. Letter in reply to the letter to the editor of Harte JV and Mykytiv V with the title "A panhaemocytometric approach to COVID-19: a retrospective study on the importance of monocyte and neutrophil population data". Clin Chem Lab Med 2021;59:e173-4. [Crossref] [PubMed]
- BC-6800 Plus Auto Hematology Analyser. Operator’s manual. Mindray Bio-medical Electronics Co., Ltd., Shenzhen, China; 2018. Available online: https://www.mindray.com/es/products/laboratory-diagnostics/hematology/5-part-differential-analyzers/bc-6800-plus
- Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol 2020;92:424-32. [Crossref] [PubMed]
- Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med 2020;58:1131-4. [Crossref] [PubMed]
- Henriot I, Launay E, Boubaya M, et al. New parameters on the hematology analyzer XN-10 (SysmexTM) allow to distinguish childhood bacterial and viral infections. Int J Lab Hematol 2017;39:14-20. [Crossref] [PubMed]
- Urrechaga E, Bóveda O, Aguirre U, et al. Neutrophil Cell Population Data Biomarkers for Acute Bacterial Infection. J Pathol Infect Dis 2018;1:1-7. [Crossref]
- Karimi Shahri M, Niazkar HR, Rad F. COVID-19 and hematology findings based on the current evidences: A puzzle with many missing pieces. Int J Lab Hematol 2021;43:160-8. [Crossref] [PubMed]
- Urrechaga E, Ponga C, Fernández M, et al. Diagnostic potential of leukocyte differential and cell population data in prediction of COVID-19 among related viral and bacterial infections at Emergency Department. Clin Chem Lab Med 2022;60:e104-7. [Crossref] [PubMed]
- Yang AP, Liu JP, Tao WQ, et al. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol 2020;84:106504. [Crossref] [PubMed]
- Lagunas-Rangel FA. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. J Med Virol 2020;92:1733-4. [Crossref] [PubMed]
- Qin C, Zhou L, Hu Z, et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 2020;71:762-8. [Crossref] [PubMed]
- Chen Z, John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol 2020;20:529-36. [Crossref] [PubMed]
- Schiff G, Mirica M. COVID-19: making the right diagnosis. Diagnosis (Berl) 2020;7:377-80. [Crossref] [PubMed]
Cite this article as: Urrechaga E, España PP, Uranga A, Fernández M, Haider RZ, Garcia de Guadiana L, Aguirre U. Leukocyte differential and cell population data in the evaluation of SARS-CoV-2 and other infections. J Lab Precis Med 2023;8:2.